3DeeCellTracker
 3DeeCellTracker copied to clipboard
3DeeCellTracker copied to clipboard
A python algorithm for tracking cells in 3D time lapse images.
3DeeCellTracker
3DeeCellTracker is a deep-learning based pipeline for tracking cells in 3D time-lapse images of deforming/moving organs (eLife, 2021).
Updates:
3DeeCellTracker v1.0.0 has been released
- Fixed some bugs in v0.5
Installation
To install 3DeeCellTracker, please follow the instructions below:
Note: We have tested the installation and the tracking programs in two environments:
- (Local) Ubuntu 20.04; NVIDIA GeForce RTX 3080Ti; Tensorflow 2.5.0
- (Google Colab) Tensorflow 2.12.0 (You need to upload your data for tracking)
Prerequisites
- A computer with an NVIDIA GPU that supports CUDA.
- Anaconda or Miniconda installed.
- TensorFlow 2.x installed.
Steps
-
Create a new conda environment and activate it by running the following commands in your terminal:
$ conda create -n track python=3.8 pip $ conda activate track -
Install TensorFlow 2.x by following the instructions provided in the TensorFlow installation guide.
-
Install the 3DeeCellTracker package by running the following command in your terminal:
$ pip install 3DeeCellTracker==1.0.0 -
Once the installation is complete, you can start using 3DeeCellTracker for your 3D cell tracking tasks within the Jupyter notebooks provided in the GitHub repository.
If you encounter any issues or have any questions, please refer to the project's documentation or raise an issue in the GitHub repository.
Quick Start
- Important: Please use the notebooks here if you installed version 1.0.0 from PyPI. If you installed from the source code available at this repository, use the notebooks provided below.
To learn how to track cells using 3DeeCellTracker, please refer to the following notebooks for examples. We recommend using StarDist for segmentation, as we have optimized the StarDist-based tracking programs for more convenient and quick cell tracking. Users can also use the old way with 3D U-Net.
-
Train a custom deep neural network for segmenting cells in new optical conditions:
- Train 3D StarDist (notebook with results)
- Train 3D U-Net (clear notebook)
- Train 3D U-Net (results)
-
Track cells in deforming organs:
- Single mode + StarDist (notebook with results)
- Single mode + UNet (clear notebook)
- single mode + UNet (results)
-
Track cells in freely moving animals:
- Ensemble mode + StarDist (notebook with results)
- Ensemble mode + UNet (clear notebook)
- Ensemble mode + UNet (results)
-
(Optional) Train FFN with custom data:
- Use coordinates in a .csv file (notebook with results)
- Use manually corrected segmentation saved as label images (notebook with results)
The data and model files for demonstrating above notebooks can be downloaded here:
Protocols
Here are two protocols for using 3DeeCellTracker:
-
Version 1.0.0
Wen, C. (2024). Deep Learning-Based Cell Tracking in Deforming Organs and Moving Animals. In: Wuelfing, C., Murphy, R.F. (eds) Imaging Cell Signaling. Methods in Molecular Biology, vol 2800. Humana, New York, NY. DOI: 10.1007/978-1-0716-3834-7_14
-
Version 0.4
Wen, C. and Kimura, K.D. (2022). Tracking Moving Cells in 3D Time Lapse Images Using 3DeeCellTracker. Bio-protocol 12(4): e4319. DOI: 10.21769/BioProtoc.4319
Frequently Reported Issue and Solution (for v0.4)
Multiple users have reported encountering a ValueError of shape mismatch when running the tracker.match() function.
After investigation, it was found that the issue resulted from an incorrect setting of siz_xyz,
which should be set to the dimensions of the 3D image as (height, width, depth).
Video Tutorials (for v0.4)
We have made tutorials explaining how to use our software. See links below (videos in Youtube):
Tutorial 1: Install 3DeeCellTracker and train the 3D U-Net
Tutorial 2: Tracking cells by 3DeeCellTracker
Tutorial 3: Annotate cells for training 3D U-Net
Tutorial 4: Manually correct the cell segmentation
How it works
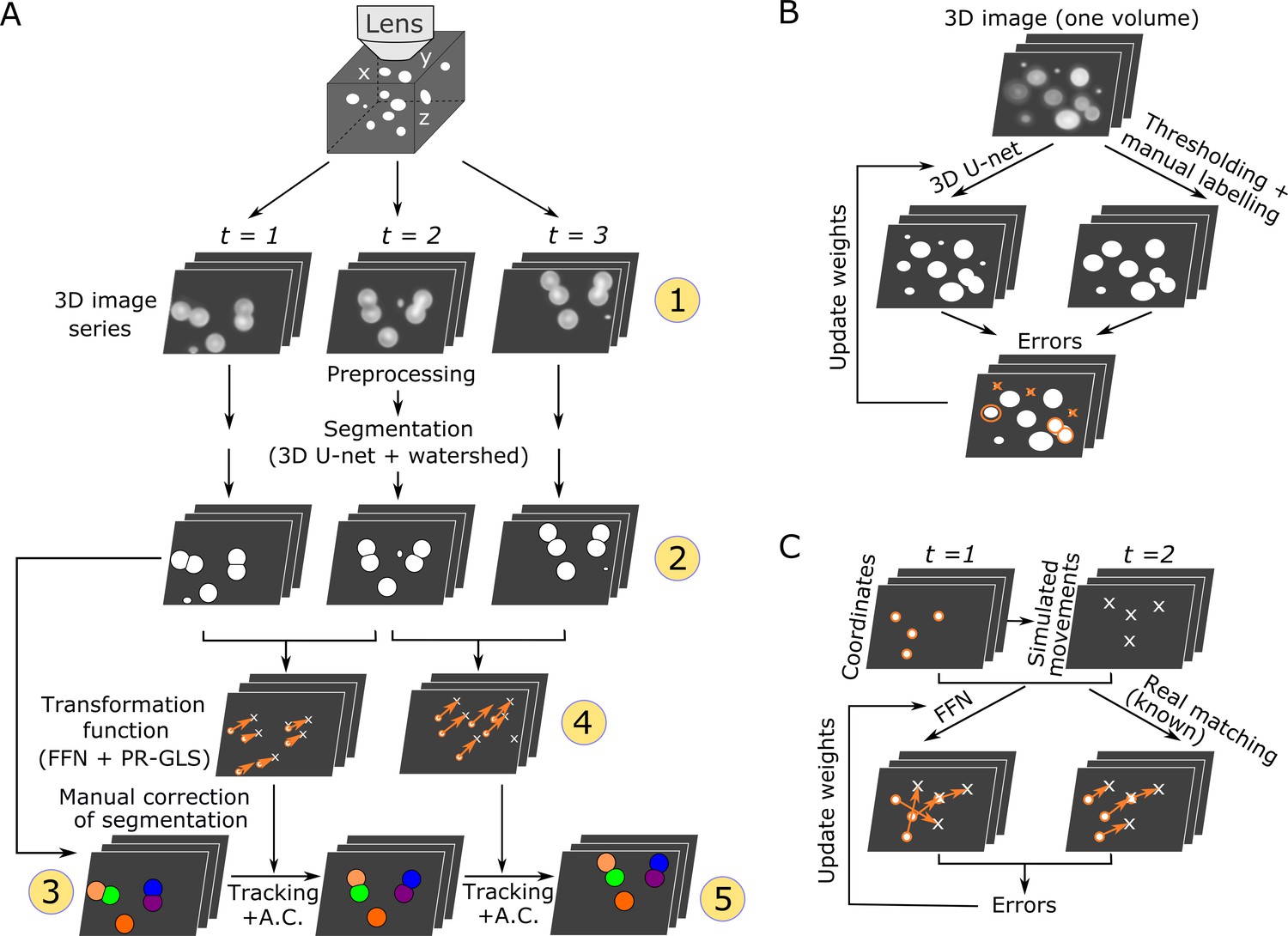
We designed this pipeline for segmenting and tracking cells in 3D + T images in deforming organs. The methods have been explained in Wen et al. bioRxiv 2018 and in Wen et al. eLife, 2021. The original programs used in eLife 2021 was contained in the "Deprecated_programs" folder.
Overall procedures of our method (Wen et al. eLife, 2021–Figure 1)
Examples of tracking results (Wen et al. eLife, 2021–Videos)
| Neurons in a ‘straightened’ freely moving worm |
Cardiac cells in a zebrafish larva | Cells in a 3D tumor spheriod |
|---|---|---|
 |
 |
 |
Citation
If you used this package in your research, please cite our paper:
- Chentao Wen, Takuya Miura, Venkatakaushik Voleti, Kazushi Yamaguchi, Motosuke Tsutsumi, Kei Yamamoto, Kohei Otomo, Yukako Fujie, Takayuki Teramoto, Takeshi Ishihara, Kazuhiro Aoki, Tomomi Nemoto, Elizabeth MC Hillman, Koutarou D Kimura (2021) 3DeeCellTracker, a deep learning-based pipeline for segmenting and tracking cells in 3D time lapse images eLife 10:e59187
Depending on the segmentation method you used (StarDist3D or U-Net3D), you may also cite either of following papers:
-
Martin Weigert, Uwe Schmidt, Robert Haase, Ko Sugawara, and Gene Myers. Star-convex Polyhedra for 3D Object Detection and Segmentation in Microscopy. The IEEE Winter Conference on Applications of Computer Vision (WACV), Snowmass Village, Colorado, March 2020
-
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O. (2016). 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M., Unal, G., Wells, W. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science(), vol 9901. Springer, Cham.
Acknowledgements
We wish to thank JetBrains for supporting this project with free open source Pycharm license.
