












Click here for more information
Table of Contents
- Introduction
- Development process
- Recent updates
- Features
- Maintenance
- Try your first PAMI program
- Evaluation
- Reading Material
- License
- Documentation
- Background
- Getting Help
- Discussion and Development
- Contribution to PAMI
- Tutorials
- Real-World Case Studies
Introduction
PAttern MIning (PAMI) is a Python library containing several algorithms to discover user interest-based patterns in a wide-spectrum of datasets across multiple computing platforms. Useful links to utilize the services of this library were provided below:
-
Youtube tutorial https://www.youtube.com/playlist?list=PLKP768gjVJmDer6MajaLbwtfC9ULVuaCZ
-
Tutorials (Notebooks) https://github.com/UdayLab/PAMI/tree/main/notebooks
-
User manual https://udaylab.github.io/PAMI/manuals/index.html
-
Coders manual https://udaylab.github.io/PAMI/codersManual/index.html
-
Code documentation https://pami-1.readthedocs.io
-
Datasets https://u-aizu.ac.jp/~udayrage/datasets.html
-
Discussions on PAMI usage https://github.com/UdayLab/PAMI/discussions
-
Report issues https://github.com/UdayLab/PAMI/issues
Process Flow Chart

Recent Updates
- Version 2023.07.07: New algorithms: cuApriroi, cuAprioriBit, cuEclat, cuEclatBit, gPPMiner, cuGPFMiner, FPStream, HUPMS, SHUPGrowth New codes to generate synthetic databases
- Version 2023.06.20: Fuzzy Partial Periodic, Periodic Patterns in High Utility, Code Documentation, help() function Update
- Version 2023.03.01: prefixSpan and SPADE
Total number of algorithms: 83
Features
- ✅ Well-tested and production-ready
- 🔋 Highly optimized to our best effort, light-weight, and energy-efficient
- 👀 Proper code documentation
- 🍼 Ample examples of using various algorithms at ./notebooks folder
- 🤖 Works with AI libraries such as TensorFlow, PyTorch, and sklearn.
- ⚡️ Supports Cuda and PySpark
- 🖥️ Operating System Independence
- 🔬 Knowledge discovery in static data and streams
- 🐎 Snappy
- 🐻 Ease of use
Maintenance
Installation
-
Installing basic pami package (recommended)
pip install pami
-
Installing pami package in a GPU machine that supports CUDA
pip install 'pami[gpu]'
-
Installing pami package in a distributed network environment supporting Spark
pip install 'pami[spark]'
-
Installing pami package for developing purpose
pip install 'pami[dev]'
-
Installing complete Library of pami
pip install 'pami[all]'
Upgradation
pip install --upgrade pami
Uninstallation
pip uninstall pami
Information
pip show pami
Try your first PAMI program
$ python
# first import pami
from PAMI.frequentPattern.basic import FPGrowth as alg
fileURL = "https://u-aizu.ac.jp/~udayrage/datasets/transactionalDatabases/Transactional_T10I4D100K.csv"
minSup=300
obj = alg.FPGrowth(iFile=fileURL, minSup=minSup, sep='\t')
obj.startMine()
obj.save('frequentPatternsAtMinSupCount300.txt')
frequentPatternsDF= obj.getPatternsAsDataFrame()
print('Total No of patterns: ' + str(len(frequentPatternsDF))) #print the total number of patterns
print('Runtime: ' + str(obj.getRuntime())) #measure the runtime
print('Memory (RSS): ' + str(obj.getMemoryRSS()))
print('Memory (USS): ' + str(obj.getMemoryUSS()))
Output:
Frequent patterns were generated successfully using frequentPatternGrowth algorithm
Total No of patterns: 4540
Runtime: 8.749667644500732
Memory (RSS): 522911744
Memory (USS): 475353088
Evaluation:
- we compared three different Python libraries such as PAMI, mlxtend and efficient-apriori for Apriori.
- (Transactional_T10I4D100K.csv)is a transactional database downloaded from PAMI and
used as an input file for all libraries.
- Minimum support values and seperator are also same.
- The performance of the Apriori algorithm is shown in the graphical results below:
-
Comparing the Patterns Generated by different Python libraries for the Apriori algorithm:
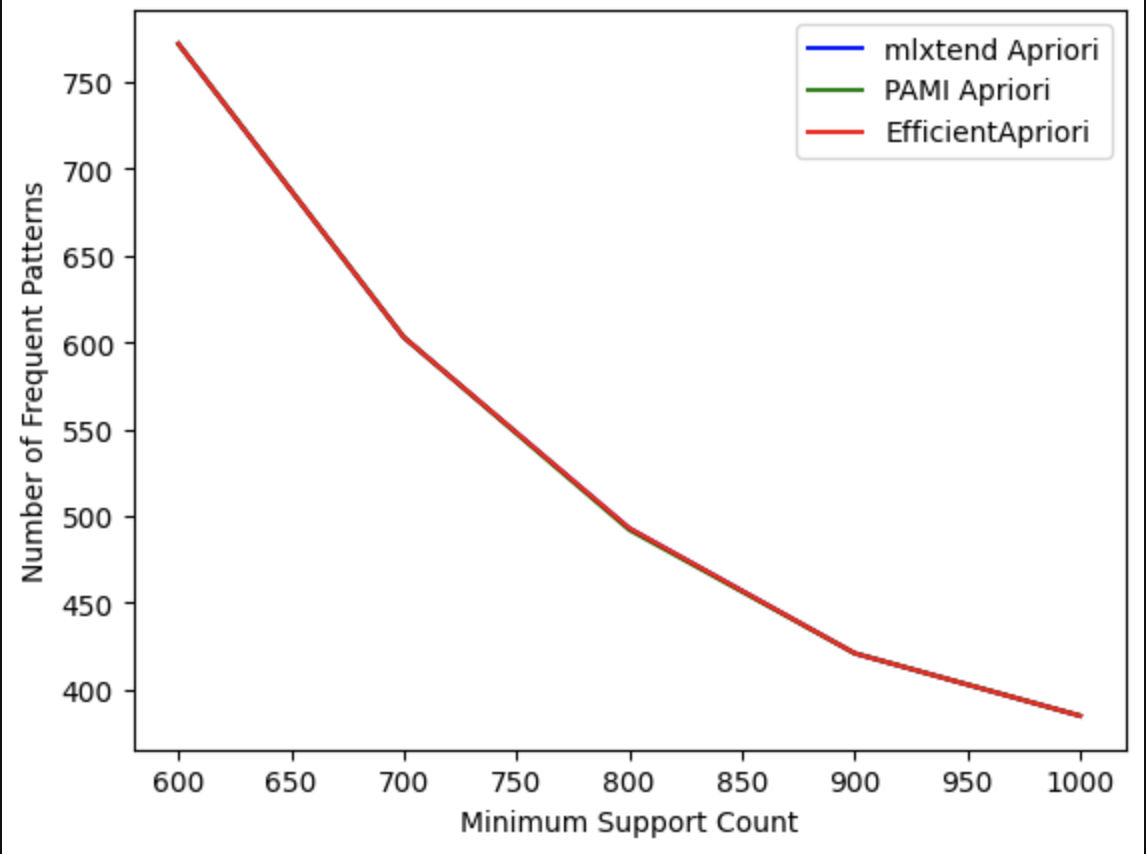
-
Evaluating the Runtime of the Apriori algorithm across different Python libraries:
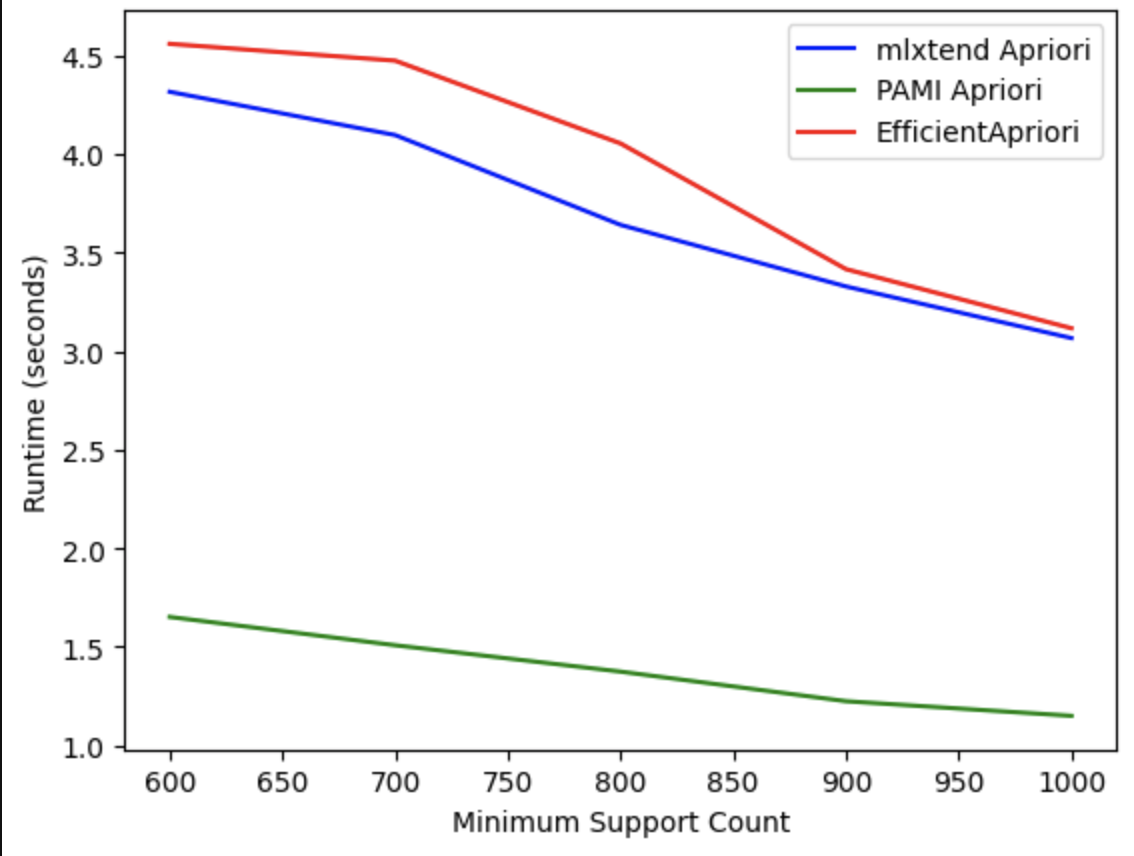
-
Comparing the Memory Consumption of the Apriori algorithm across different Python libraries:
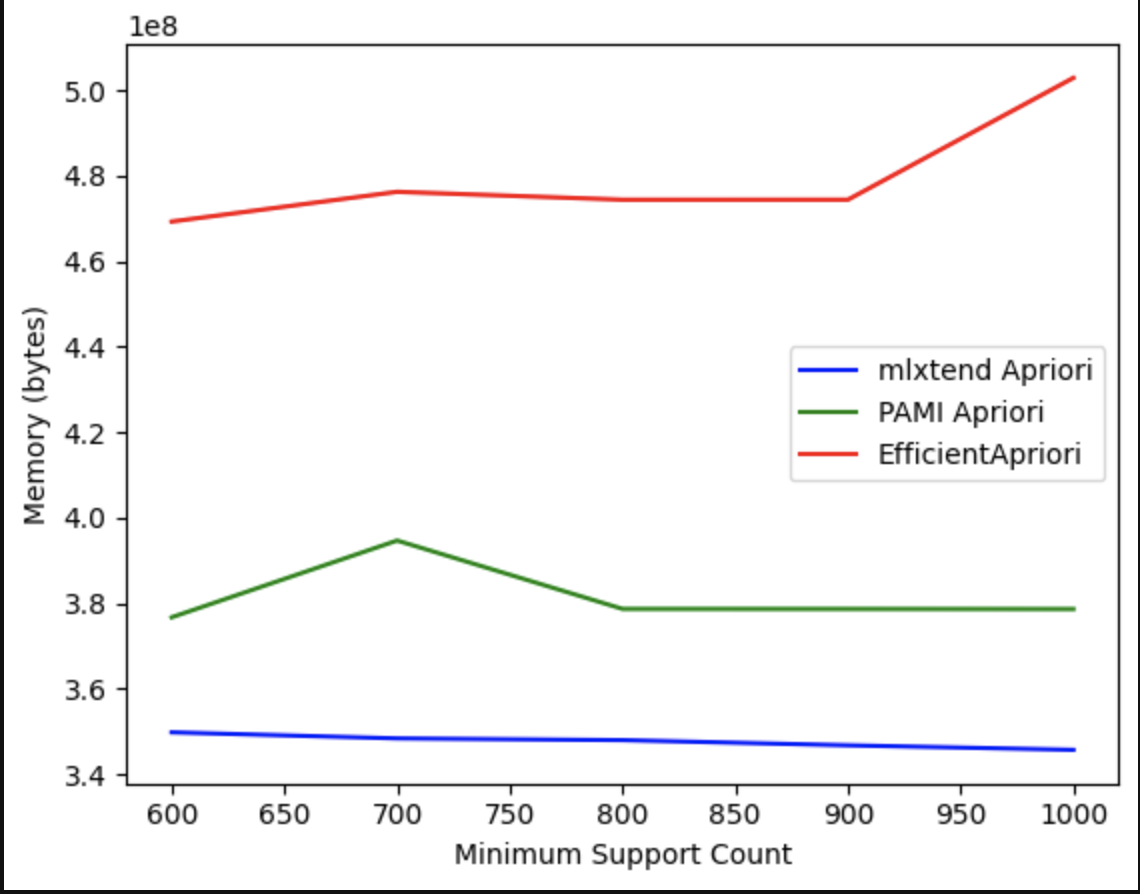
For more information, we have uploaded the evaluation file in two formats:
Reading Material
For more examples, refer this YouTube link YouTube
License

Documentation
The official documentation is hosted on PAMI.
Background
The idea and motivation to develop PAMI was from Kitsuregawa Lab at the University of Tokyo. Work on PAMI started at University of Aizu in 2020 and
has been under active development since then.
Getting Help
For any queries, the best place to go to is Github Issues GithubIssues.
Discussion and Development
In our GitHub repository, the primary platform for discussing development-related matters is the university lab. We encourage our team members and contributors to utilize this platform for a wide range of discussions, including bug reports, feature requests, design decisions, and implementation details.
Contribution to PAMI
We invite and encourage all community members to contribute, report bugs, fix bugs, enhance documentation, propose improvements, and share their creative ideas.
Tutorials
0. Association Rule Mining
| Basic |
Confidence  |
Lift  |
Leverage  |
1. Pattern mining in binary transactional databases
1.1. Frequent pattern mining: Sample
| Basic |
Closed |
Maximal |
Top-k |
CUDA |
pyspark |
Apriori  |
CHARM  |
maxFP-growth  |
FAE  |
cudaAprioriGCT |
parallelApriori  |
FP-growth  |
|
|
|
cudaAprioriTID |
parallelFPGrowth  |
ECLAT  |
|
|
|
cudaEclatGCT |
parallelECLAT  |
ECLAT-bitSet  |
|
|
|
|
|
ECLAT-diffset  |
|
|
|
|
|
1.2. Relative frequent pattern mining: Sample
| Basic |
RSFP-growth  |
1.3. Frequent pattern with multiple minimum support: Sample
| Basic |
CFPGrowth  |
CFPGrowth++  |
1.4. Correlated pattern mining: Sample
| Basic |
CoMine  |
CoMine++  |
1.5. Fault-tolerant frequent pattern mining (under development)
| Basic |
FTApriori  |
FTFPGrowth (under development)  |
1.6. Coverage pattern mining (under development)
| Basic |
CMine  |
CMine++  |
2. Pattern mining in binary temporal databases
2.1. Periodic-frequent pattern mining: Sample
| Basic |
Closed |
Maximal |
Top-K |
PFP-growth  |
CPFP  |
maxPF-growth  |
kPFPMiner  |
PFP-growth++  |
|
Topk-PFP  |
|
PS-growth  |
|
|
|
PFP-ECLAT  |
|
|
|
PFPM-Compliments  |
|
|
|
2.2. Local periodic pattern mining: Sample
| Basic |
LPPGrowth (under development)  |
LPPMBreadth (under development)  |
LPPMDepth (under development)  |
2.3. Partial periodic-frequent pattern mining: Sample
| Basic |
GPF-growth  |
PPF-DFS  |
GPPF-DFS  |
2.4. Partial periodic pattern mining: Sample
| Basic |
Closed |
Maximal |
topK |
CUDA |
3P-growth  |
3P-close  |
max3P-growth  |
topK-3P growth  |
cuGPPMiner (under development)  |
3P-ECLAT  |
|
|
|
gPPMiner (under development)  |
G3P-Growth  |
|
|
|
|
2.5. Periodic correlated pattern mining: Sample
| Basic |
EPCP-growth  |
2.6. Stable periodic pattern mining: Sample
| Basic |
TopK |
SPP-growth  |
TSPIN  |
SPP-ECLAT  |
|
2.7. Recurring pattern mining: Sample
| Basic |
RPgrowth  |
3. Mining patterns from binary Geo-referenced (or spatiotemporal) databases
3.1. Geo-referenced frequent pattern mining: Sample
| Basic |
spatialECLAT  |
FSP-growth  |
3.2. Geo-referenced periodic frequent pattern mining: Sample
| Basic |
GPFPMiner  |
PFS-ECLAT  |
ST-ECLAT  |
3.3. Geo-referenced partial periodic pattern mining:Sample
| Basic |
STECLAT  |
4. Mining patterns from Utility (or non-binary) databases
4.1. High utility pattern mining: Sample
| Basic |
EFIM  |
HMiner  |
UPGrowth  |
4.2. High utility frequent pattern mining: Sample
| Basic |
HUFIM  |
4.3. High utility geo-referenced frequent pattern mining: Sample
| Basic |
SHUFIM  |
4.4. High utility spatial pattern mining: Sample
| Basic |
topk |
HDSHIM  |
TKSHUIM  |
SHUIM  |
|
4.5. Relative High utility pattern mining: Sample
| Basic |
RHUIM  |
4.6. Weighted frequent pattern mining: Sample
| Basic |
WFIM  |
4.7. Weighted frequent regular pattern mining: Sample
| Basic |
WFRIMiner  |
4.8. Weighted frequent neighbourhood pattern mining: Sample
5. Mining patterns from fuzzy transactional/temporal/geo-referenced databases
5.1. Fuzzy Frequent pattern mining: Sample
| Basic |
FFI-Miner  |
5.2. Fuzzy correlated pattern mining: Sample
| Basic |
FCP-growth  |
5.3. Fuzzy geo-referenced frequent pattern mining: Sample
| Basic |
FFSP-Miner  |
5.4. Fuzzy periodic frequent pattern mining: Sample
| Basic |
FPFP-Miner  |
5.5. Fuzzy geo-referenced periodic frequent pattern mining: Sample
| Basic |
FGPFP-Miner (under development)  |
6. Mining patterns from uncertain transactional/temporal/geo-referenced databases
6.1. Uncertain frequent pattern mining: Sample
| Basic |
top-k |
PUF  |
TUFP |
TubeP  |
|
TubeS  |
|
| UVEclat |
|
6.2. Uncertain periodic frequent pattern mining: Sample
| Basic |
UPFP-growth  |
UPFP-growth++  |
6.3. Uncertain Weighted frequent pattern mining: Sample
| Basic |
WUFIM  |
7. Mining patterns from sequence databases
7.1. Sequence frequent pattern mining: Sample
| Basic |
SPADE  |
PrefixSpan  |
7.2. Geo-referenced Frequent Sequence Pattern mining
| Basic |
GFSP-Miner (under development)  |
8. Mining patterns from multiple timeseries databases
8.1. Partial periodic pattern mining (under development)
| Basic |
PP-Growth (under development)  |
9. Mining interesting patterns from Streams
- Frequent pattern mining
- High utility pattern mining
10. Mining patterns from contiguous character sequences (E.g., DNA, Genome, and Game sequences)
10.1. Contiguous Frequent Patterns
| Basic |
PositionMining  |
11. Mining patterns from Graphs
11.1. Frequent sub-graph mining
| Basic |
topk |
Gspan  |
TKG  |
12. Additional Features
12.1. Creation of synthetic databases
| Database type |
Transactional database  |
Temporal database  |
| Utility database (coming soon) |
12.2. Converting a dataframe into a specific database type
| Approaches |
| Dense dataframe to databases (coming soon) |
| Sparse dataframe to databases (coming soon) |
12.3. Gathering the statistical details of a database
| Approaches |
| Transactional database (coming soon) |
| Temporal database (coming soon) |
| Utility database (coming soon) |
12.4. Generating Latex code for the experimental results
| Approaches |
| Latex code (coming soon) |
Real World Case Studies
- Air pollution analytics

Go to Top
 PAMI copied to clipboard
PAMI copied to clipboard












