s-jSDM
 s-jSDM copied to clipboard
s-jSDM copied to clipboard
Easy Model Residuals
Hi, first of all thanks for the great package. Loving this as I can now run community models at such a large geographic scale compared to other methods such as HMSC, it's made some infeasible projects suddenly feasible for my PhD!
I wanted to know if there was any easy way implemented to get the model residuals? I've tried accounting for spatial autocorrelation in my communities/models with a trend surface parameter but want to compare a model with and without this term to see the effect and further explore whether a DNN or spatial eigenvectors are worth running.
Dan
Hi @dansmi-hub,
Unfortunately, we haven't implemented a resid/residual method yet. However, you can calculate it yourself. Stand. Pearson residuals:
com = simulate_SDM(env = 3L, species = 7L, sites = 100L, correlation = FALSE)
model = sjSDM(Y = com$response,env = com$env_weights, iter = 50L)
p = predict(model)
# Calculate residuals
residuals_sjsdm = (com$response-p)/sqrt(p*(1-p))
Compare it to glm:
m = glm(com$response[,1]~com$env_weights, family = binomial("probit"))
plot(resid(m, type = "pearson"), residuals_sjsdm[,1])

Residual checks
At the moment we do not support community residual checks (there is an ongoing discussion about how to do this, https://github.com/florianhartig/DHARMa/issues/189), but you can still use the DHARMa package to check the residuals of individual species (so you don't have to calculate the residuals yourself):
- Simulate data
set.seed(42)
W = matrix(c(5, 0.5), 2, 12)
E = matrix(runif(1000*2,-1,1), 1000, 2)
Raw = (E[,1,drop=FALSE]**2)%*%W[1,,drop=FALSE] + mvtnorm::rmvnorm(1000, sigma = cov2cor(rWishart(1, 100, Sigma = diag(1., 12))[,,1]))
Y = Raw
Y = ifelse(Y > 0, 1, 0)
- Fit two models (one with the wrong functional form)
model1 = sjSDM(Y = Y,env = linear(data.frame(E), formula = ~X1), ,iter = 100L)
model2 = sjSDM(Y = Y,env = linear(data.frame(E), formula = ~I(X1^2)), ,iter = 100L)
- Simulate from the model
sp = 1
pred1 = predict(model1)[,sp]
sim1 = sapply(1:100, function(i) rbinom(1000, 1, prob = pred1))
pred2 = predict(model2)[,sp]
sim2 = sapply(1:100, function(i) rbinom(1000, 1, prob = pred2))
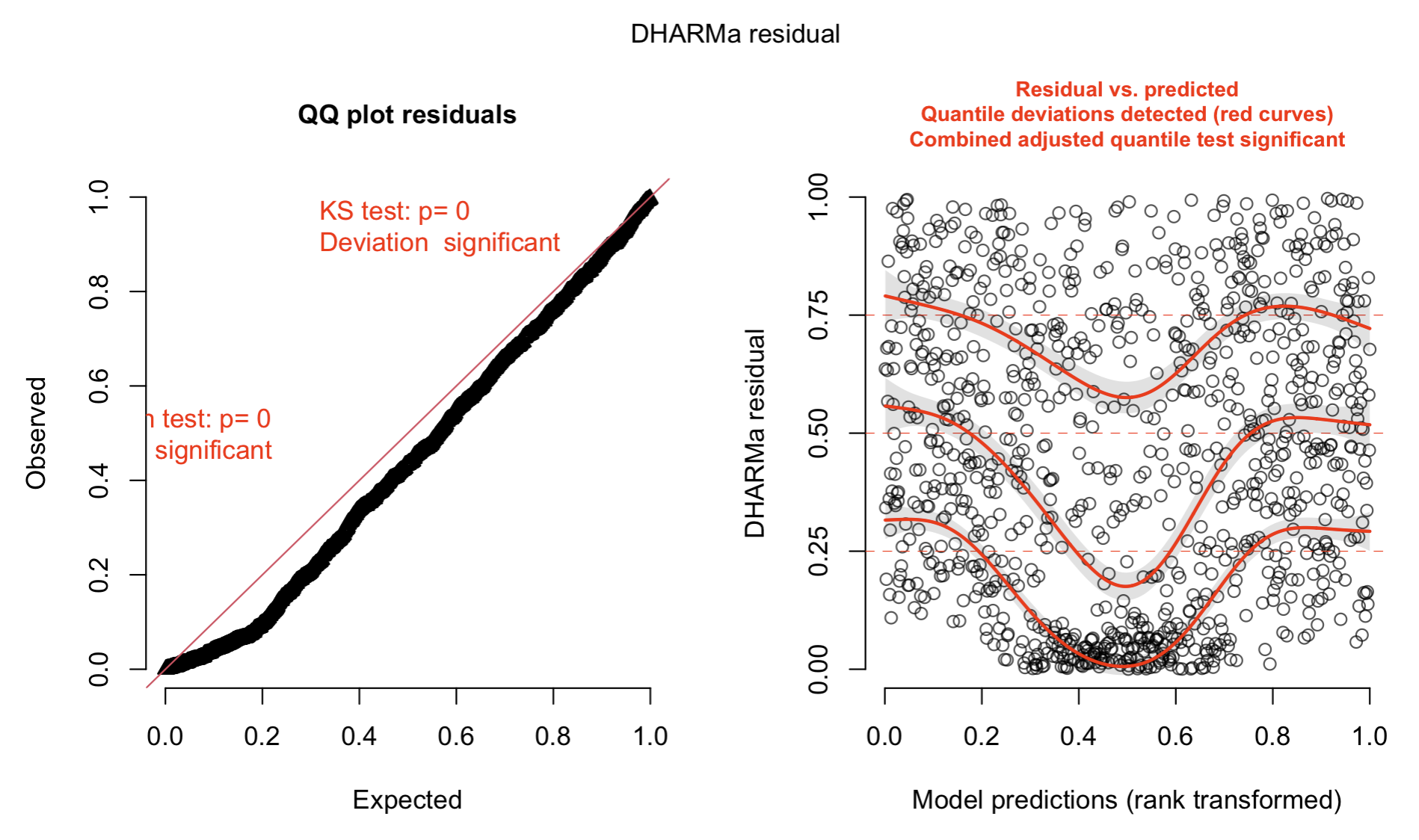
- Residual checks for one species with the DHARMa package
library(DHARMa)
res1 = DHARMa::createDHARMa(simulatedResponse = sim1,
observedResponse = Y[,sp],
fittedPredictedResponse = as.vector(predict(model1)[,sp]))
plot(res1)
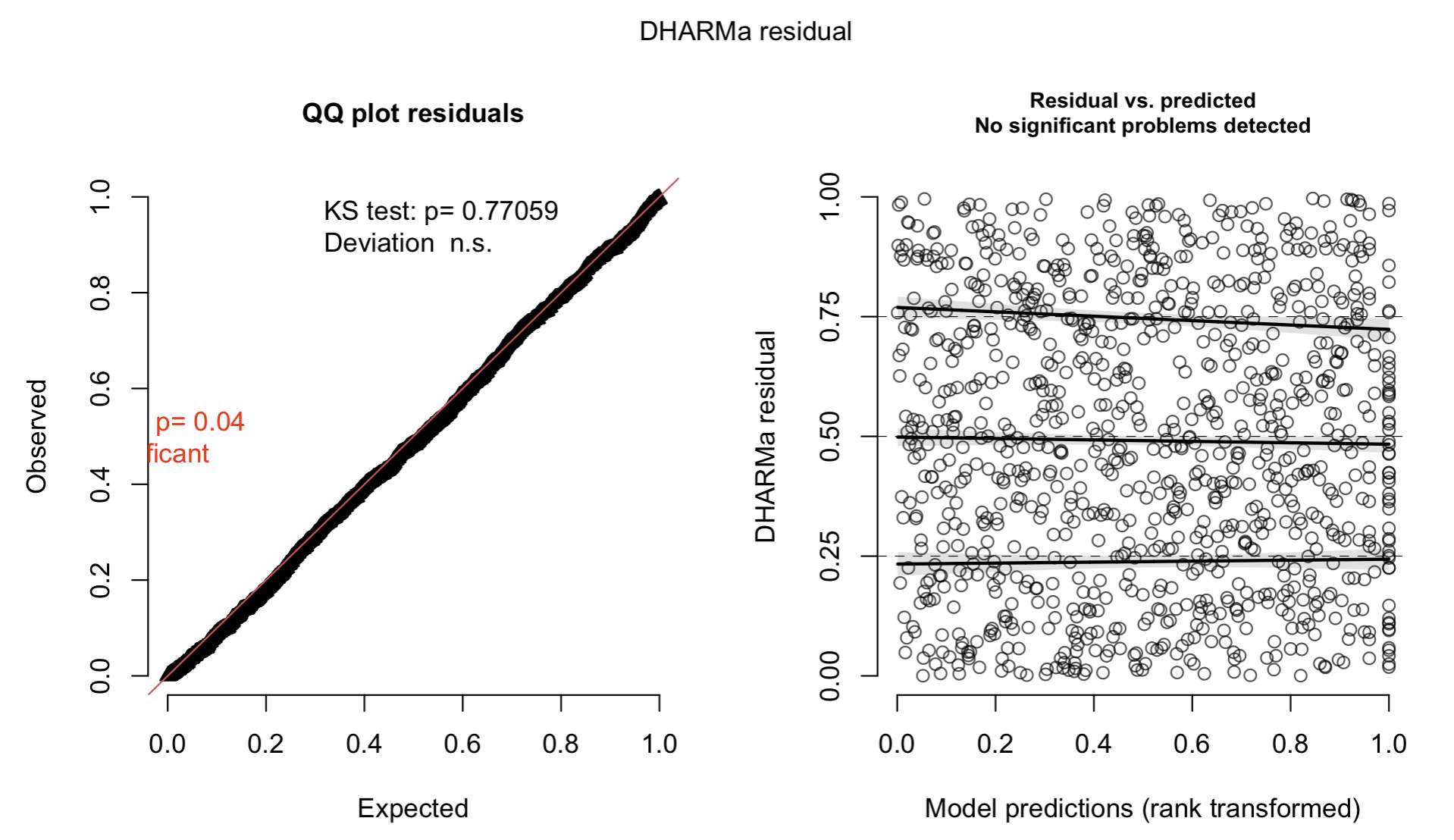
res2 = DHARMa::createDHARMa(simulatedResponse = sim2,
observedResponse = as.vector(Y[,sp]),
fittedPredictedResponse = as.vector(predict(model2)[,sp]))
plot(res2)