In Python Geometricalnfo.get_size() returns sizes in reverse order than shape for STIR.ImageData
It's incredibly confusing that img.get_geometrical_info().get_size() return a tuple that is in the reverse order of img.as_array().shape (which is luckily the same as img.get_dimensions().
Presumably get_spacing() also returns reverse order (i.e. opposite of img.voxel_sizes()). I haven't checked.
I don't know how it is in C++ nor MATLAB.
Changing this might have some consequences elsewhere and needs to be done with caution I guess.
We also have some interesting name differences in these functions of course. This is at the moment fortuitous as they return something else, but it's not nice.
thanks for fixing the Python side @evgueni-ovtchinnikov. However, is the underlying C++ code not problematic either? And what about get_spacing etc?
Could you check if any of these are used in the demos/SIRF-exercises?
C++: in STIR, images dimensions order is zyx, in ISMRMRD xyz, so a synergistic code cannot avoid confusion (cf. MATLAB vs Python array dimensions).
I did change get_offset and get_spacing too.
And I did check they are not used anywhere except one place in test_pReg.py, where dimensions, however, are all equal.
C++: in STIR, images dimensions order is zyx, in ISMRMRD xyz, so a synergistic code cannot avoid confusion (cf. MATLAB vs Python array dimensions).
I would like to avoid terms like xyz as much as possible. With image data types that can store objects in various orientations, and patients that can be put in the scanner in various orientation, that terminology becomes rather hard to stick to 😉 .
In fact, this is now starting to give me a headache.
get_offsetshould be in LPS (it's a coordinate that in a coordinate system that is fixed to the patient, independent of how the data stores it).get_sizes()to me has to reflect the array sizes of the "native" array object (i.e. in Pythonas_array().shape, in MATLABas_array().size()). In C++, this isn't so obvious, as SIRF actually doesn't directly store the array (it just access it from STIR/Nifty/ISMRMRD/whatever). I guess we do have the iterators that walk sequentially through the data, so it still makes sense to tell people what the sizes of an nD-array would be, filled that way. (which I guess means in C-order).get_spacing()to me has to be in the same order asget_sizes(): it gives you mm spacing in you walk along a particular dimension
get_sizes() and get_spacing() should therefore depend on order (with first element of get_spacing() determined as norm(matrix. [1,0,0,1]), and therefore potentially on the engine.
@DANAJK might have an opinion.
I did change
get_offsetandget_spacingtoo.
I did the following.
- take an Interfile image and convert it to NIfTI with STIR (no other program can do this). (I used
stir_mathfor that.) - read it in SIRF with both STIR and
Reg. Here's what I get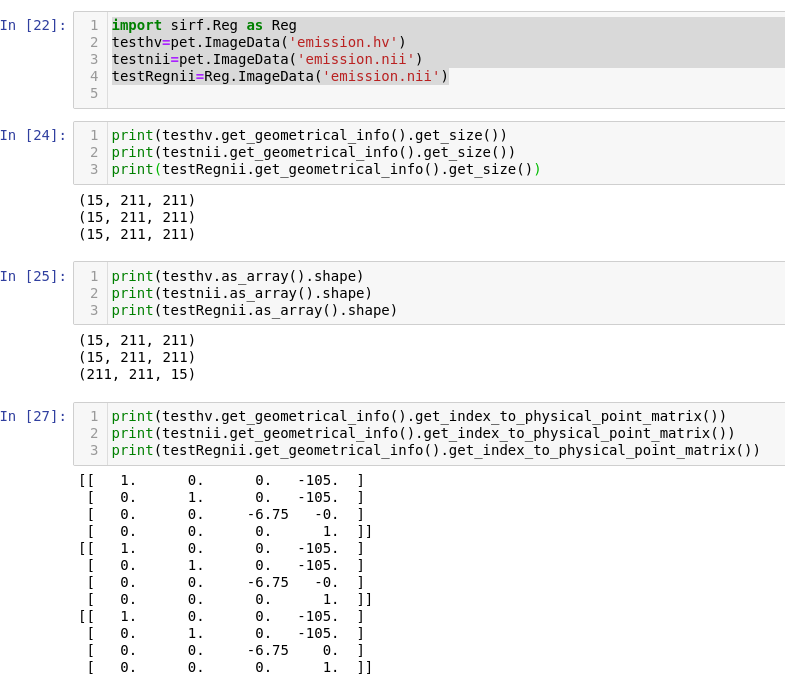 i.e. order of the underlying data is different but the direction matrices (and size/offset/index->physical matrix) are all the same. Only one of these can be correct of course.
i.e. order of the underlying data is different but the direction matrices (and size/offset/index->physical matrix) are all the same. Only one of these can be correct of course.
I also checked in https://github.com/SyneRBI/SIRF-Exercises/pull/111 that resampling worked ok with both STIRImageData and Reg.NiftiImageData. After all this, that's surprising!
The relevant lines are https://github.com/SyneRBI/SIRF/blob/28317c13d9599b86da26b85ca6556479f17db87b/src/xSTIR/cSTIR/stir_data_containers.cpp#L615-L629 where @rijobro reverts the order from STIR, and https://github.com/SyneRBI/SIRF/blob/28317c13d9599b86da26b85ca6556479f17db87b/src/xSTIR/cSTIR/stir_data_containers.cpp#L633-L643 which also does various reorderings.
The Reg case makes most sense to me - the 3rd array dimension has shape value of 15 which I assume corresponds to slice, and the corresponding 3rd column of the matrix has the largest absolute size (6.75) which is also likely to be slice.
Note that NIfTI essentially says files should be in Fortran format (first array index varies most rapidly). This is taken care of in nibabel, and, I assume NiftyReg.
The Reg case makes most sense to me - the 3rd array dimension has shape value of 15 which I assume corresponds to slice, and the corresponding 3rd column of the matrix has the largest absolute size (6.75) which is also likely to be slice.
agreed
Note that NIfTI essentially says files should be in Fortran format
really? I'm amazed.
In any case, whatever we decide to read data in, the geometrical_info has to adjust obviously.
From the NIfTI header:
N.B.: The i index varies most rapidly, j index next, k index slowest.
Thus, voxel (i,j,k) is stored starting at location
(i + j*dim[1] + k*dim[1]*dim[2]) * (bitpix/8)
into the dataset array.
From nibabel:
array_from_file
order{‘F’, ‘C’} string
order in which to write data. Default is ‘F’ (fortran order).
https://github.com/SyneRBI/SIRF/pull/969 should revert everything to normal for Nifti, and therefore changes STIR. I had to disable the sirf.STIR.ImageData tests for geometrical_info therefore.
Merging #969 closed this automatically, but it didn't actually fix the issue...
I'm afraid I'm running out of time today for an update before the Dev meeting.
But I'm narrowing in and may have found the issue.
We've tried to be tricky here and reverse STIR's reversal, so that in SIRF we wouldn't have something like SPL instead of LPS (trying to avoid x, y, z notation - but note I mean gantry not patient)
https://github.com/SyneRBI/SIRF/blob/699b12ceb2e8d74d804c2a3620875d8a969a7843/src/xSTIR/cSTIR/stir_data_containers.cpp#L615-L629
But we interface with the data directly:
https://github.com/SyneRBI/SIRF/blob/699b12ceb2e8d74d804c2a3620875d8a969a7843/src/xSTIR/cSTIR/cstir.cpp#L1327-L1349
So I think we are basically always swapping the definitions of the first and last axes for GeomInfo. David's image is isotropic in the LS direction, so it comes out correct. Most images aren't
David's image is isotropic in the LS direction
aha. Indeed. I see that get_info() after reading as STIR.ImageData
Offset: (-180.82, -21.1697, 384.01)
Spacing: (0.925926, 5.73606, 0.925926)
Size: (432, 30, 432)
Direction matrix:
1, 0, 0
0, 1, 0
0, 0, -1
so the fact that it worked with @DANAJK's data is an accident, and it is a real bug. That at least is less confusing
I don't understand these comments fully, but I just want to clarify that the data has pixel widths and heights that are the same, but the slice thickness is different. If you want a dataset where the L and S pixel sizes are different, use the axial slices (OBJECT_phantom_T2W_TSE_Tra_17_1.nii). Remember that Spacing should be in the image coordinate system, not LPS. If Spacing were in LPS, then the values would change as you angulate a slice.
let's first create non-isotropic test data
as I mentioned above "If you want a dataset where the L and S pixel sizes are different, use the axial slices (OBJECT_phantom_T2W_TSE_Tra_17_1.nii)."