Is whitespace allowed in CURIEs?
Reading through the spec: https://www.w3.org/TR/2010/NOTE-curie-20101216/
I don't find an obvious mention whether whitespace is permitted in a CURIE, but whitespace separated list of CURIEs appears to be a thing:
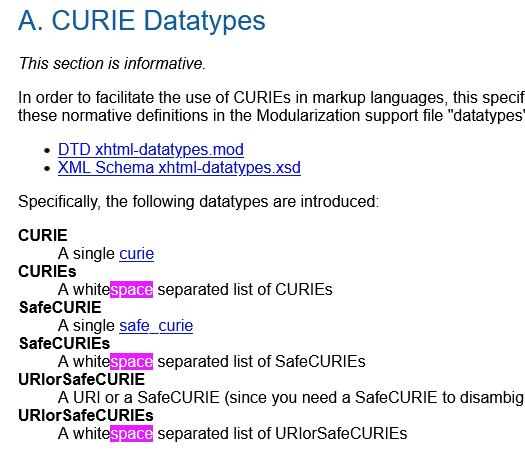
suggesting to me that CURIEs should/must(?) not contain whitespace?
But it appears we do have these? https://github.com/RTXteam/RTX/issues/1233#issuecomment-802544001
Is that a problem?
I concur. This seems like it might be a bug.
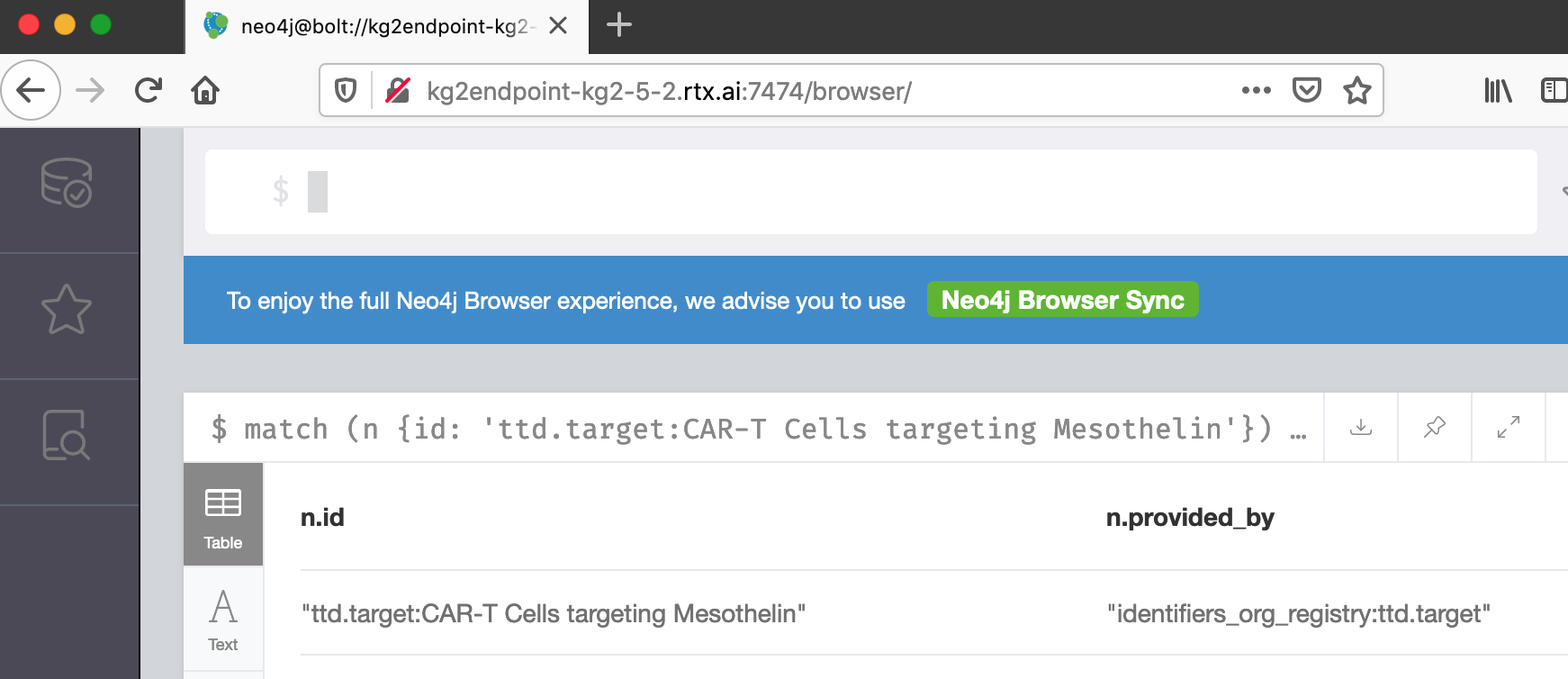
This code
match (n {id: 'ttd.target:CAR-T Cells targeting Mesothelin'}) return n.id, n.provided_by;
produces:
"ttd.target:CAR-T Cells targeting Mesothelin" | "identifiers_org_registry:ttd.target"
-- | --
on KG2.5.2:
we probably want to look upstream of 105 of dgidb_tsv_to_kg_json.py:
https://github.com/RTXteam/RTX/blob/2300b9f9e985cd6ccbf127a195bcd73544e54fdc/code/kg2/dgidb_tsv_to_kg_json.py#L105
to where subject_curie_id is getting set.
Thank you @edeutsch and @amykglen for bringing this to my attention; very helpful linking the CURIE spec.
I think I found the offending line in dgidb's interactions.tsv:
MSLN Mesothelin 10232 TTD CAR-T cells targeting mesothelin CAR-T cells targeting mesothelin
Based on it's entry on TTD's website (see here):

it looks like the correct ID for this node is T87108. The term T87108 is not present in interactions.tsv.
This may be way out in left field, but this feels similar to the issue in RTXteam/RTX#449/#434.
In general, the whitespace nodes seem to be created within the following lines: https://github.com/RTXteam/RTX/blob/397967a413a4bc998a3fc20f4a6d8fa79db67c62/code/kg2/dgidb_tsv_to_kg_json.py#L99-L110
I verified that the all of the whitespace containing curie ids come from ttd.target using the following cypher:
match (n) where n.id contains " " return distinct n.provided_by, count(*)
| n.provided_by | count(*) |
|---|---|
| "identifiers_org_registry:ttd.target" | 3550 |
As such, I went ahead and replaced the whitespace in these node ids with underscores. (thanks Erica, for tracking down where it's coming from!)
I did the same for the IRI field, though it's still not resolving. Looking at https://registry.identifiers.org/registry/ttd.target, we would need a different format of TTD identifiers that are NOT included in the interactions TSV to make them resolvable. Though the example (https://identifiers.org/ttd.target:TTDS00056) provided in the identifiers.org link doesn't appear to be working either .....
Looking at https://registry.identifiers.org/registry/ttd.target, we would need a different format of TTD identifiers that are NOT included in the interactions TSV to make them resolvable. Though the example (https://identifiers.org/ttd.target:TTDS00056) provided in the identifiers.org link doesn't appear to be working either .....
I reached out to identifiers.org about this a few days back and they haven't gotten back to me yet. There appears to be another website providing TTD information, but I'm unsure if it is legitimate. The source of TTD listed in their publication no longer resolves.
In its current state, is it worth keeping the TTD information in KG2? Since the identifiers are incorrect, is it helpful?
This is fixed in kg2.6.0, but I'm going to leave it open to discuss the inclusion of TTD in kg2!
Adding the sar-look label to weigh in on @ericawood 's above question
DELETED
@saramsey If you look at the TTD identifiers currently in KG2.6.0, you'll notice that the identifiers are just the names of the nodes. The TTD nodes are created in dgidb_tsv_to_kg_json.py:
https://github.com/RTXteam/RTX/blob/cce89beffe7aebdd9f0585fe448b3ad0db2d444d/code/kg2/dgidb_tsv_to_kg_json.py#L103-L107
match (n) where split(n.id, ':')[0]='ttd.drug' or split(n.id, ':')[0]='ttd.target' return n.id, n.name limit 50
| n.id | n.name |
|---|---|
| "ttd.target:Fondaparinux_sodium" | "Fondaparinux sodium" |
| "ttd.target:PMID27454349-Compound-95" | "PMID27454349-Compound-95" |
| "ttd.target:Pancreatic_cancer_vaccine" | "Pancreatic cancer vaccine" |
| "ttd.target:PMID27977313-Compound-48" | "PMID27977313-Compound-48" |
| "ttd.target:Imidazo[5,1-c]pyrido[2,3-e][1,2,4]triazine_derivative_6" | "Imidazo[5,1-c]pyrido[2,3-e][1,2,4]triazine derivative 6" |
| "ttd.target:JX-929" | "JX-929" |
| "ttd.target:PMID29473428-Compound-50" | "PMID29473428-Compound-50" |
| "ttd.target:US8470836,_5" | "US8470836, 5" |
| "ttd.target:EB-101" | "EB-101" |
| "ttd.target:PMID25666693-Compound-128" | "PMID25666693-Compound-128" |
| "ttd.target:Tetra-hydro-naphthalene_derivative_2" | "Tetra-hydro-naphthalene derivative 2" |
| "ttd.target:ATD_transdermal_gel" | "ATD transdermal gel" |
| "ttd.target:SCHEMBL17766424" | "SCHEMBL17766424" |
| "ttd.target:HouttuynoidA" | "HouttuynoidA" |
| "ttd.target:Peptide_analog_45" | "Peptide analog 45" |
| "ttd.target:Dihydropyrimidinone_derivative_3" | "Dihydropyrimidinone derivative 3" |
| "ttd.target:Indazoletriazolephenyl_derivative_1" | "Indazoletriazolephenyl derivative 1" |
| "ttd.target:Pyrrolidine_derivative_10" | "Pyrrolidine derivative 10" |
| "ttd.target:LOXO-292" | "LOXO-292" |
| "ttd.target:PMID29671355-Compound-18" | "PMID29671355-Compound-18" |
| "ttd.target:US9682983,_1" | "US9682983, 1" |
| "ttd.target:Aryl_urea_derivative_2" | "Aryl urea derivative 2" |
| "ttd.target:PMID25522065-Compound-36" | "PMID25522065-Compound-36" |
| "ttd.target:Pyrimidine_derivative_1" | "Pyrimidine derivative 1" |
| "ttd.target:Heterocycle-containing_compound_1" | "Heterocycle-containing compound 1" |
| "ttd.target:PMID27454349-Compound-94" | "PMID27454349-Compound-94" |
| "ttd.target:Alkynyl-substituted_pyrimidinyl-pyrrole_derivative_1" | "Alkynyl-substituted pyrimidinyl-pyrrole derivative 1" |
| "ttd.target:Amidopyrazole_derivative_5" | "Amidopyrazole derivative 5" |
| "ttd.target:Ketoheterocycle_derivative_3" | "Ketoheterocycle derivative 3" |
| "ttd.target:TGWOOAA" | "TGWOOAA" |
| "ttd.target:PMID25522065-Compound-34" | "PMID25522065-Compound-34" |
| "ttd.target:MM-151" | "MM-151" |
| "ttd.target:Isoquinoline_derivative_5" | "Isoquinoline derivative 5" |
| "ttd.target:TBC-3711" | "TBC-3711" |
| "ttd.target:2-pyrazinone_derivative_7" | "2-pyrazinone derivative 7" |
| "ttd.target:Imidazo[1,2-b]pyridazine_acetamide_derivative_7" | "Imidazo[1,2-b]pyridazine acetamide derivative 7" |
| "ttd.target:Pyrazole_derivative_67" | "Pyrazole derivative 67" |
| "ttd.target:Pyrrolidinyl_urea_derivative_13" | "Pyrrolidinyl urea derivative 13" |
| "ttd.target:Carboxyamidotriazole_orotate" | "Carboxyamidotriazole orotate" |
| "ttd.target:Mycophenolic_acid/nucleotide_derivative_1" | "Mycophenolic acid/nucleotide derivative 1" |
| "ttd.target:CL-316,243" | "CL-316,243" |
| "ttd.target:PMID26004420-Compound-WO2014126944A" | "PMID26004420-Compound-WO2014126944A" |
| "ttd.target:Tricyclic_compound_7" | "Tricyclic compound 7" |
| "ttd.target:Imidazole_derivative_7" | "Imidazole derivative 7" |
| "ttd.target:Sulfonamide_derivative_16" | "Sulfonamide derivative 16" |
| "ttd.target:Descartes-08" | "Descartes-08" |
| "ttd.target:Cyclopropyl-spiro_piperidine_derivative_4" | "Cyclopropyl-spiro piperidine derivative 4" |
| "ttd.target:PMID26394986-Compound-Figure17" | "PMID26394986-Compound-Figure17" |
| "ttd.target:Thiazole_derivative_2" | "Thiazole derivative 2" |
| "ttd.target:TAK-020" | "TAK-020" |
Thank you @ericawood, that helps. OK, so my previous comment was based on a misunderstanding on my part. It sounds like the DGIdb ETL process is only creating TTD nodes for TTD drugs not targets, right?
In that case, I think perhaps there isn't anything we can do with the DGIdb cross-references to TTD that would result in a resolvable IRI for the TTD drug node, what do you think, @ericawood and @kvarforl ?
I don't see where DGIdb is getting the node "names" for TTD drugs from. They are not perfect matches to a field in the TTD data dump files, as far as I can tell.
What about the TTD cross-references from DrugBank, are those also problematic? see https://github.com/RTXteam/RTX/blob/cce89beffe7aebdd9f0585fe448b3ad0db2d444d/code/kg2/drugbank_xml_to_kg_json.py#L237
Thank you @ericawood, that helps. OK, so my previous comment was based on a misunderstanding on my part. It sounds like the DGIdb ETL process is only creating TTD nodes for TTD drugs not targets, right?
I think that's backwards:
https://github.com/RTXteam/RTX/blob/cce89beffe7aebdd9f0585fe448b3ad0db2d444d/code/kg2/dgidb_tsv_to_kg_json.py#L32-L33
It appears that DGIdb is only creating nodes for TTD targets.
HOWEVER, that is not the main issue. The bigger issue is that the IDs it is creating are NOT actual TTD IDs.
Also, DGIdb's strange node "names" are not just a TTD issue. That has been part of the issue with RTXteam/RTX#449. DGIdb's node "names" don't exactly match the NCIT dump either.
What about the TTD cross-references from DrugBank, are those also not useful?
Those aren't helpful because there aren't actual TTD nodes in KG2.
@ericawood The identifiers you listed above, while they may have the CURIE prefix "ttd.target", look to be drugs to me. I'm pretty sure they are not drug targets in actuality (though I admit the CURIE prefix makes it seem that way).
@ericawood The identifiers you listed above, while they may have the CURIE prefix "ttd.target", look to be drugs to me. I'm pretty sure they are not drug targets in actuality (though I admit the CURIE prefix makes it seem that way).
Got it, thank you for your scientific understanding! Should we switch the prefix?
@ericawood The identifiers you listed above, while they may have the CURIE prefix "ttd.target", look to be drugs to me. I'm pretty sure they are not drug targets in actuality (though I admit the CURIE prefix makes it seem that way).
Got it, thank you for your scientific understanding! Should we switch the prefix?
Yes, we should use the CURIE prefix ttd.drug. We also need to update curies-to-urls.yaml so that the prefix ttd.drug maps to the URL: http://db.idrblab.net/ttd/search/ttd/drug?search_api_fulltext=. And instead of changing spaces to underscores in the drug name, maybe we should change them to "%20". Then an ID like ttd.drug:Carboxyamidotriazole%20orotate should (uh, please fact check my reasoning here) resolve to
http://db.idrblab.net/ttd/search/ttd/drug?search_api_fulltext=Carboxyamidotriazole%20orotate
which actually works (yes it is kind of yucky, but perfect is the enemy of good).
So, identifiers.org is nifty, but DGIdb doesn't give us the nice numeric TTD identifiers that we would need, to actually reference a TTD drug (or target) via identifiers.org. The fault here is squarely with DGIdb. But on the other hand, good edges with publication info are hard to come by.
Will other sources link to TTD properly with the identifiers in this form?
I contacted the identifiers.org help desk a while back about their TTD identifiers not resolving. This was their response:
Dear Erica Wood,
thank you for pointing out this problem, we have requested the new URLs from the resource providers.
Best regards,
Henning
In KG2.8.3, running
match (n) where n.id contains " " return distinct n.provided_by, count(*)
returns no results, but the TTD identifiers are still do not resolve:
match (n) where split(n.id, ':')[0]='ttd.drug' or split(n.id, ':')[0]='ttd.target' return n.id, n.name, n.iri limit 50
| n.id | n.name | n.iri |
|---|---|---|
| "ttd.target:AVP-13358" | "AVP-13358" | "https://identifiers.org/ttd.target:AVP-13358" |
| "ttd.target:PMID29338548-Compound-40" | "PMID29338548-Compound-40" | "https://identifiers.org/ttd.target:PMID29338548-Compound-40" |
| "ttd.target:PMID28270021-Compound-WO2010077680_109" | "PMID28270021-Compound-WO2010077680 109" | "https://identifiers.org/ttd.target:PMID28270021-Compound-WO2010077680_109" |
| "ttd.target:BMS-986165" | "BMS-986165" | "https://identifiers.org/ttd.target:BMS-986165" |
| "ttd.target:MV-CEA" | "MV-CEA" | "https://identifiers.org/ttd.target:MV-CEA" |
| "ttd.target:Actimab-M" | "Actimab-M" | "https://identifiers.org/ttd.target:Actimab-M" |
| "ttd.target:Imidazo_pyridine_derivative_4" | "Imidazo pyridine derivative 4" | "https://identifiers.org/ttd.target:Imidazo_pyridine_derivative_4" |
| "ttd.target:AphanamgrandiolA" | "AphanamgrandiolA" | "https://identifiers.org/ttd.target:AphanamgrandiolA" |
| "ttd.target:BI_655066" | "BI 655066" | "https://identifiers.org/ttd.target:BI_655066" |
| "ttd.target:JNJ-54728518" | "JNJ-54728518" | "https://identifiers.org/ttd.target:JNJ-54728518" |
| "ttd.target:Insulin_degludec" | "Insulin degludec" | "https://identifiers.org/ttd.target:Insulin_degludec" |
| "ttd.target:HER-2/HER-1_vaccine" | "HER-2/HER-1 vaccine" | "https://identifiers.org/ttd.target:HER-2/HER-1_vaccine" |
| "ttd.target:Aminoazetidine_derivative_3" | "Aminoazetidine derivative 3" | "https://identifiers.org/ttd.target:Aminoazetidine_derivative_3" |
| "ttd.target:Peptide_analog_44" | "Peptide analog 44" | "https://identifiers.org/ttd.target:Peptide_analog_44" |
| "ttd.target:Belerofon" | "Belerofon" | "https://identifiers.org/ttd.target:Belerofon" |
| "ttd.target:P11187" | "P11187" | "https://identifiers.org/ttd.target:P11187" |
| "ttd.target:AMG_579" | "AMG 579" | "https://identifiers.org/ttd.target:AMG_579" |
| "ttd.target:PMID28766366-Compound-Scheme9EHT5372" | "PMID28766366-Compound-Scheme9EHT5372" | "https://identifiers.org/ttd.target:PMID28766366-Compound-Scheme9EHT5372" |
| "ttd.target:GSK1070916A" | "GSK1070916A" | "https://identifiers.org/ttd.target:GSK1070916A" |
| "ttd.target:PMID28870136-Compound-43" | "PMID28870136-Compound-43" | "https://identifiers.org/ttd.target:PMID28870136-Compound-43" |
| "ttd.target:Peptide_analog_70" | "Peptide analog 70" | "https://identifiers.org/ttd.target:Peptide_analog_70" |
| "ttd.target:VLB-01" | "VLB-01" | "https://identifiers.org/ttd.target:VLB-01" |
| "ttd.target:XEN007" | "XEN007" | "https://identifiers.org/ttd.target:XEN007" |
| "ttd.target:MC-4_agonist" | "MC-4 agonist" | "https://identifiers.org/ttd.target:MC-4_agonist" |
| "ttd.target:Quinazoline_derivative_15" | "Quinazoline derivative 15" | "https://identifiers.org/ttd.target:Quinazoline_derivative_15" |
| "ttd.target:PMID29338548-Compound-31" | "PMID29338548-Compound-31" | "https://identifiers.org/ttd.target:PMID29338548-Compound-31" |
| "ttd.target:Carbamide_derivative_11" | "Carbamide derivative 11" | "https://identifiers.org/ttd.target:Carbamide_derivative_11" |
| "ttd.target:Fused_aryl_carbocycle_derivative_1" | "Fused aryl carbocycle derivative 1" | "https://identifiers.org/ttd.target:Fused_aryl_carbocycle_derivative_1" |
| "ttd.target:Quinoline_carboxamide_derivative_9" | "Quinoline carboxamide derivative 9" | "https://identifiers.org/ttd.target:Quinoline_carboxamide_derivative_9" |
| "ttd.target:Cyclohexyl_carbamate_derivative_5" | "Cyclohexyl carbamate derivative 5" | "https://identifiers.org/ttd.target:Cyclohexyl_carbamate_derivative_5" |
| "ttd.target:Anti-CD19-CAR_vector-transduced_T_cells" | "Anti-CD19-CAR vector-transduced T cells" | "https://identifiers.org/ttd.target:Anti-CD19-CAR_vector-transduced_T_cells" |
| "ttd.target:Pyrrolo-pyridinone_derivative_1" | "Pyrrolo-pyridinone derivative 1" | "https://identifiers.org/ttd.target:Pyrrolo-pyridinone_derivative_1" |
| "ttd.target:Tolamba" | "Tolamba" | "https://identifiers.org/ttd.target:Tolamba" |
| "ttd.target:PMID30185082-Compound-14" | "PMID30185082-Compound-14" | "https://identifiers.org/ttd.target:PMID30185082-Compound-14" |
| "ttd.target:HF-0299" | "HF-0299" | "https://identifiers.org/ttd.target:HF-0299" |
| "ttd.target:PMID27215781-Compound-13" | "PMID27215781-Compound-13" | "https://identifiers.org/ttd.target:PMID27215781-Compound-13" |
| "ttd.target:Novaferon" | "Novaferon" | "https://identifiers.org/ttd.target:Novaferon" |
| "ttd.target:NsG-0202" | "NsG-0202" | "https://identifiers.org/ttd.target:NsG-0202" |
| "ttd.target:Erythrityl_Tetranitrate" | "Erythrityl Tetranitrate" | "https://identifiers.org/ttd.target:Erythrityl_Tetranitrate" |
| "ttd.target:PMID26293650-Compound-34" | "PMID26293650-Compound-34" | "https://identifiers.org/ttd.target:PMID26293650-Compound-34" |
| "ttd.target:CAR-T_cells_targeting_MucI" | "CAR-T cells targeting MucI" | "https://identifiers.org/ttd.target:CAR-T_cells_targeting_MucI" |
| "ttd.target:AZD1419" | "AZD1419" | "https://identifiers.org/ttd.target:AZD1419" |
| "ttd.target:Anti-HER3/EGFR_DAF" | "Anti-HER3/EGFR DAF" | "https://identifiers.org/ttd.target:Anti-HER3/EGFR_DAF" |
| "ttd.target:Macrolactam_derivative_4" | "Macrolactam derivative 4" | "https://identifiers.org/ttd.target:Macrolactam_derivative_4" |
| "ttd.target:Maleimides_derivative_1" | "Maleimides derivative 1" | "https://identifiers.org/ttd.target:Maleimides_derivative_1" |
| "ttd.target:BR-4628" | "BR-4628" | "https://identifiers.org/ttd.target:BR-4628" |
| "ttd.target:3-substituted-2-furancarboxylic_acid_hydrazide_derivative_5" | "3-substituted-2-furancarboxylic acid hydrazide derivative 5" | "https://identifiers.org/ttd.target:3-substituted-2-furancarboxylic_acid_hydrazide_derivative_5" |
| "ttd.target:GSK2646264" | "GSK2646264" | "https://identifiers.org/ttd.target:GSK2646264" |
| "ttd.target:SK-NBP601" | "SK-NBP601" | "https://identifiers.org/ttd.target:SK-NBP601" |
| "ttd.target:US8901295,_F609" | "US8901295, F609" | "https://identifiers.org/ttd.target:US8901295,_F609" |
I am unsure if we should close this out, but I'm taking the verify this fix in next kg2 build label off.
I think TTD drug CURIEs maybe should start with ttd.drug:DAP per this page (which I suppose could be out of date):
https://registry.identifiers.org/registry/ttd.drug
The ttd.drug:DAP is I guess followed by one or more digits. So perhaps the CURIE doesn't exist as a string with a DAP prefix as such in the database, but maybe it exists as an integer column in some drug table?
So maybe we can check if the TTD database has these DAP identifiers (or the integer part of the identifier) in it?
In any event, I recommend we keep this issue open for now (but to be clear, I don't think this issue needs to be fixed in the KG2.8.4pre build).