model-zoo
 model-zoo copied to clipboard
model-zoo copied to clipboard
using the original labels, instead of inverted transformed labels for evaluation
Currently quite a few bundles use the inverted version of preprocessed original labels as the reference during evaluation:
https://github.com/Project-MONAI/model-zoo/blob/987f49fe9e0f20f047a2252c1ace38bd760cfb33/models/spleen_ct_segmentation/configs/evaluate.json#L12-L21
which is different from the approach taken in the tutorial using the original label directly during evaluation (https://github.com/Project-MONAI/tutorials/blob/main/3d_segmentation/spleen_segmentation_3d.ipynb)
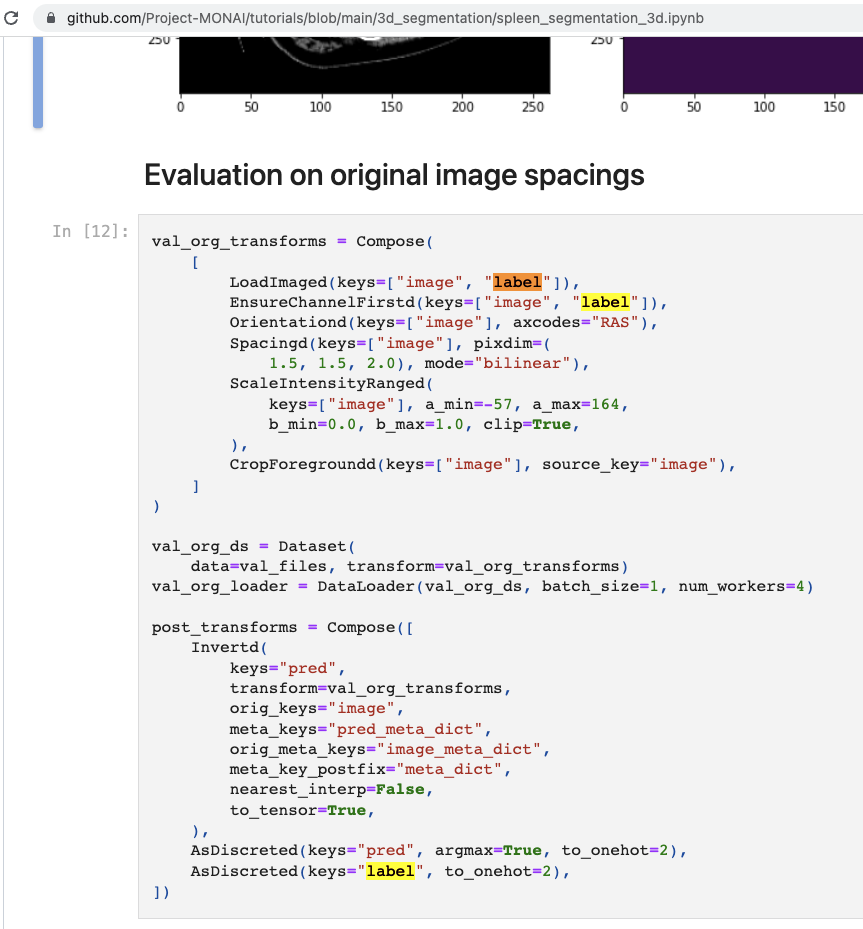
would be great to consider the standard method used in the tutorial for bundle evaluate.json
cc @holgerroth @dongyang0122
(this needs an update as well) https://github.com/Project-MONAI/tutorials/tree/main/bundle/spleen_segmentation)
It is necessary to validate without modifying the label data for (almost) all models in the model zoo.
Hi @Nic-Ma @wyli , to avoid maintaining duplicate spleen bundles in tutorial and model zoo, should we remove the one in the tutorial, and add a step in the readme file of tutorial to do "download from model zoo"?
Hi @yiheng-wang-nv ,
I think that's OK, and just put a link to the source code of spleen bundle in the model-zoo. @wyli What do you think?
Thanks.
sure, i think it'll be a separate new feature request in the tutorial repo, or is it related to this issue?
(this device='cpu' fix will be needed for Invertd https://github.com/Project-MONAI/tutorials/pull/1042 for the monai v1.1 bundles)
sure, i think it'll be a separate new feature request in the tutorial repo, or is it related to this issue?
Yes, it should be a separate new feature request in the tutorial repo, I just created one: https://github.com/Project-MONAI/tutorials/issues/1043
As for the issue of using inverted labels, may need further discussions with @Nic-Ma @daguangxu @dongyang0122 @wyli
For the issue, I checked and the following bundles need to be updated:
spleen_ct_segmentation(invert transformed labels for evaluation)endoscopic_tool_segmentation(no invert in postprocessing)pancreas_ct_dints_segmentation(invert transformed labels for evaluation)spleen_deepedit_annotation(no invert in postprocessing)swin_unetr_btcv_segmentation(invert transformed labels for evaluation)
(this
device='cpu'fix will be needed forInvertdProject-MONAI/tutorials#1042 for the monai v1.1 bundles)
Hi @wyli , I don't fully understand this requirement of device='cpu'. Would you mind explaining a bit more, for example, if we don't set it to cpu, will it break something in the spleen segmentation tutorial?
yes, it will raise this error when computing metrics: https://github.com/Project-MONAI/tutorials/issues/1041
@wyli Thanks!
For the issue, I checked and the following bundles need to be updated:
spleen_ct_segmentation(invert transformed labels for evaluation)endoscopic_tool_segmentation(no invert in postprocessing)pancreas_ct_dints_segmentation(invert transformed labels for evaluation)spleen_deepedit_annotation(no invert in postprocessing)swin_unetr_btcv_segmentation(invert transformed labels for evaluation)
Update: to prevent unexpected errors, this issue will be addressed after our next release.