Update `SobelGradients` to include `direction`
Fixes #5188
Description
This PR updates SobelGradients and SobelGradientsd to include gradient direction. It also adds unit tests.
Types of changes
- [x] Non-breaking change (fix or new feature that would not break existing functionality).
- [x] New tests added to cover the changes.
- [x] Integration tests passed locally by running
./runtests.sh -f -u --net --coverage. - [x] Quick tests passed locally by running
./runtests.sh --quick --unittests --disttests. - [x] In-line docstrings updated.
Hi @drbeh,
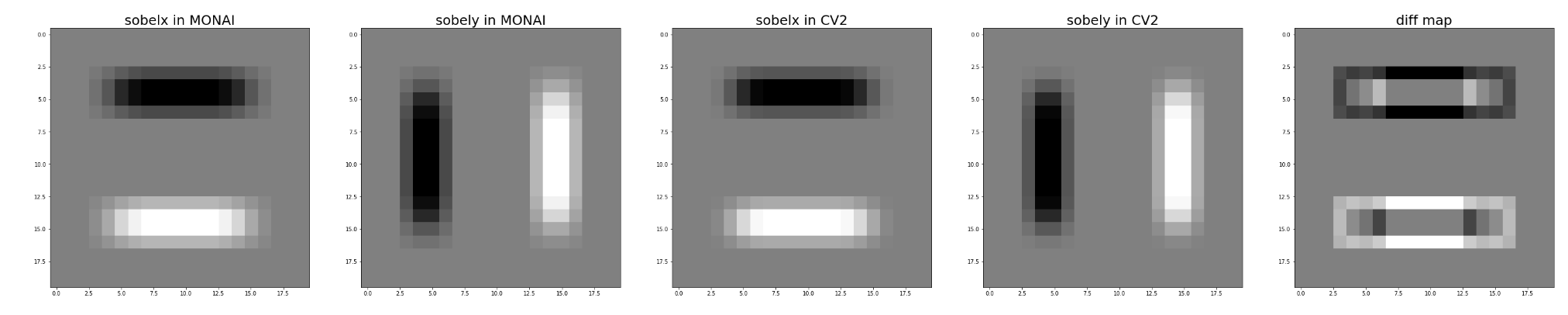
I have just added unittest in CalcualteInstanceSegmentationMap which used SobelGradients and found some different effects between cv2 and SobelGradients.
The difference between cv2 and SobelGradients also cause CalcualteInstanceSegmentationMap can't get correct instance segmentation results. So I just used cv2 now. I will withdraw my approval of this PR and please check it.
Here is my test code.
import cv2
from monai.transforms.post.array import SobelGradients
image = np.zeros([20,20])
image[5:15, 5:15] = 1
filter = SobelGradients(kernel_size=5)
mo_ret = filter(image[None]).squeeze()
cv_ret_0 = cv2.Sobel(image, cv2.CV_64F, 0, 1, ksize=5)
cv_ret_1 = cv2.Sobel(image, cv2.CV_64F, 1, 0, ksize=5)
mo_ret = 1 - (mo_ret - np.min(mo_ret)) / (np.max(mo_ret) - np.min(mo_ret))
cv_ret_0 = 1 - (cv_ret_0 - np.min(cv_ret_0)) / (np.max(cv_ret_0) - np.min(cv_ret_0))
cv_ret_1 = 1 - (cv_ret_1 - np.min(cv_ret_1)) / (np.max(cv_ret_1) - np.min(cv_ret_1))
fig, ax = plt.subplots(1,5,figsize = (50,10))
ax[0].imshow(mo_ret[0], cmap='gray')
ax[1].imshow(mo_ret[1], cmap='gray')
ax[2].imshow(cv_ret_0, cmap='gray')
ax[3].imshow(cv_ret_1, cmap='gray')
ax[4].imshow(mo_ret[0] - cv_ret_0, cmap='gray')
Thanks!
I have just added unittest in
CalcualteInstanceSegmentationMapwhich usedSobelGradientsand found some different effects between cv2 andSobelGradients
@drbeh and I were discussing this in another thread, opencv uses the separable implementation cv2.getDerivKernels(1, 0, 5), that's slightly different from what we have here. but we haven't looked into the actual segmentation outcome differences.
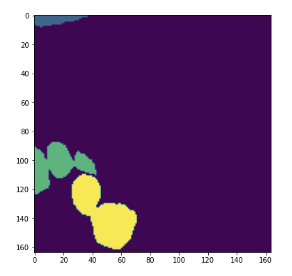
Just FYI, I showed the instance segmentation result from CalcualteInstanceSegmentationMap. The results are calculated with all the same components except for Sobel gradient.
cv2 - result
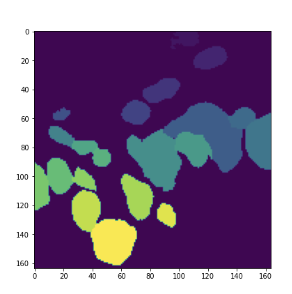 MONAI - result
MONAI - result
 Thanks!
Thanks!
Sorry - inadvertantly closed the PR
What I was trying to do was to comment that I don't recall seeing that same effect (i.e. the instance segmentation difference), so just wondering whether it really is just the sobel that is making the difference. Will look at my original code to see if I can reproduce.
Okay, so my sobel implementation, which used correlate2d from scipy.signal, predated the Monai Sobel implementation, which would explain the difference.
Hi @KumoLiu @JHancox,
As @wyli mentioned, Sobel gradients in OpenCV for higher dimensions is implemented differently but they should match exactly for a kernel size of 3x3. The main reason for our implementation is to match TIAToolbox and PathML implementation for hovernet.
@KumoLiu can you check the gradient differences with a 3x3 kernel? Also can you create the instance segmentation result from CalcualteInstanceSegmentationMap with 3x3 kernel both with opencv and monai? Thanks
Hi @drbeh, I checked the gradient differences with a 3x3 kernel.
For the faked data I generated, it seems there is no difference:
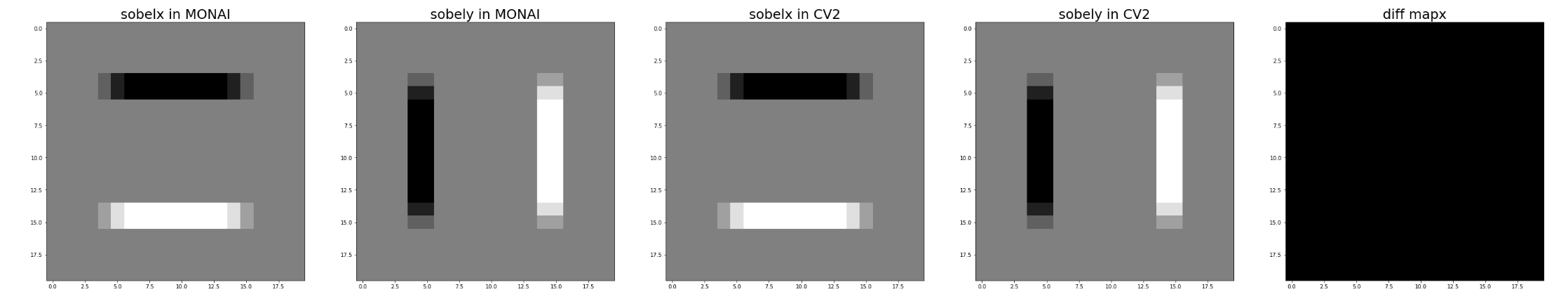 For the test data @JHancox shared with me, it seems cv2 had sharper edges:
For the test data @JHancox shared with me, it seems cv2 had sharper edges:
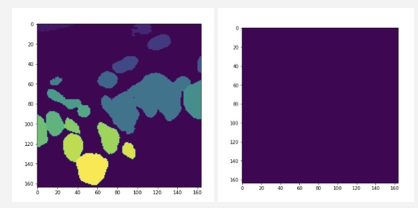
 For instance segmentation results, left is the result generated by cv2, right is generated by
For instance segmentation results, left is the result generated by cv2, right is generated bySobelGradients which generates a completely blank plot. It may be due to the threshold set here. https://github.com/Project-MONAI/tutorials/blob/156a459e95b12f0650b2cc57994f7bb8080b6cff/pathology/hovernet_nuclei_identification/post_processing.py#L142
So I also tried different thresholds, the results are listed below.
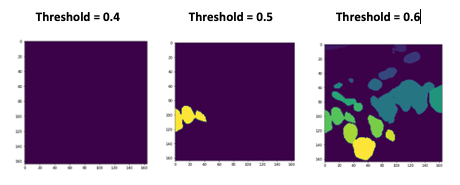 Thanks!
Thanks!
I'm sure you know but, just to be sure, HoverNet uses a sobel kernel size of 5x5 :)
Hi @KumoLiu, thank you very much for comparing the results. It seems that it is related to threshold but in fact it shouldn't. The only thing that I can think of is that maybe they are using different normalization factor for the kernel, so the output is shifted with a constant but I believe there is a normalization process happening afterwards to bring them between 0 and 1, right?
Hi @KumoLiu, thank you very much for comparing the results. It seems that it is related to threshold but in fact it shouldn't. The only thing that I can think of is that maybe they are using different normalization factor for the kernel, so the output is shifted with a constant but I believe there is a normalization process happening afterwards to bring them between 0 and 1, right?
Yes, the results I plotted here had been normalized with MinMax.
Hi @wyli, Based on our previous conversation, I have updated Sobel gradients to use separable kernels of any size and can be applied to the images with any dimension and along any given axis. Would be great if you can review. Thanks
Hi @KumoLiu, could you please run your experiment for Sobel gradients again to compare it to OpenCV implementation? thanks
Hi @drbeh, I have just tested it with kernel_size=3 on fake data and data @JHancox shared with me.
It seems we get the exact opposite of the value of OpenCV.
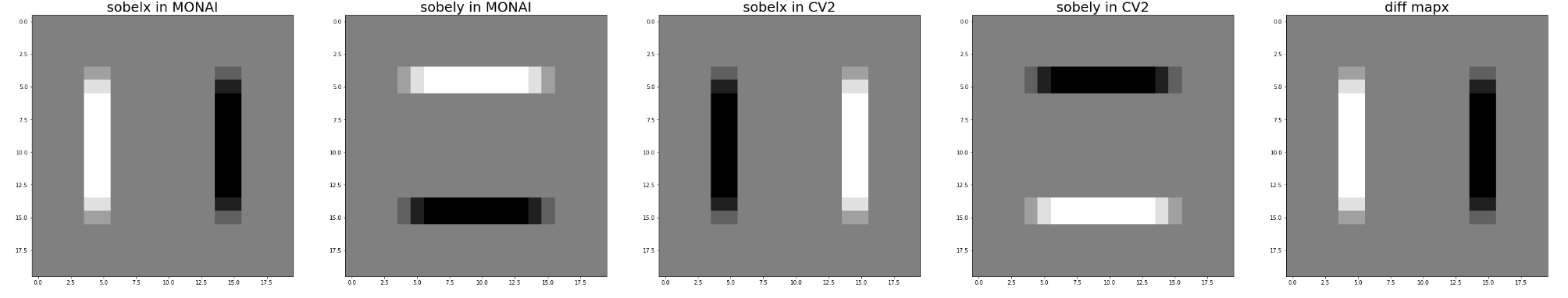
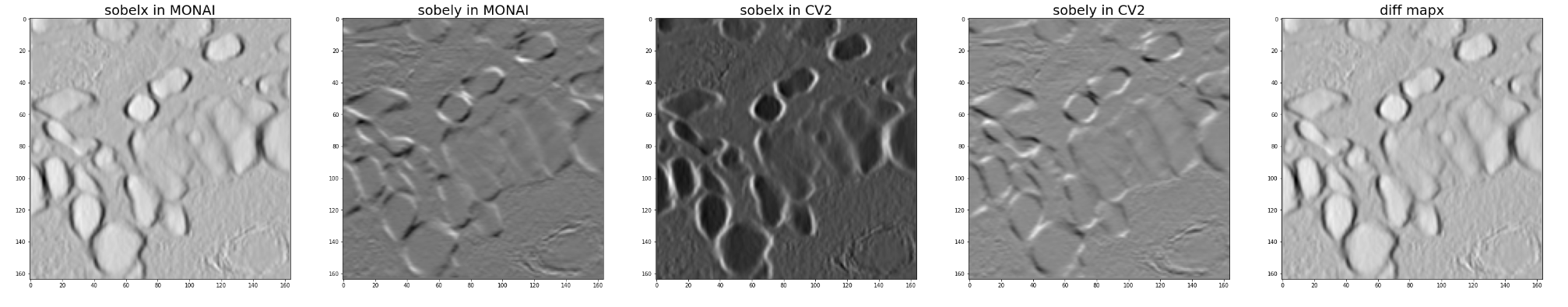
Here is my test code.
# fake data
image = np.zeros([20,20])
image[5:15, 5:15] = 1
# real data
image = hv_dir[0]
filter = SobelGradients(kernel_size=3)
mo_ret = filter(image[None]).squeeze()
cv_ret_0 = cv2.Sobel(image, cv2.CV_64F, 1, 0, ksize=3)
cv_ret_1 = cv2.Sobel(image, cv2.CV_64F, 0, 1, ksize=3)
mo_ret_0 = 1 - (mo_ret[0] - np.min(mo_ret[0])) / (np.max(mo_ret[0]) - np.min(mo_ret[0]))
mo_ret_1 = 1 - (mo_ret[1] - np.min(mo_ret[1])) / (np.max(mo_ret[1]) - np.min(mo_ret[1]))
cv_ret_0 = 1 - (cv_ret_0 - np.min(cv_ret_0)) / (np.max(cv_ret_0) - np.min(cv_ret_0))
cv_ret_1 = 1 - (cv_ret_1 - np.min(cv_ret_1)) / (np.max(cv_ret_1) - np.min(cv_ret_1))
fig, ax = plt.subplots(1,5,figsize = (50,10))
ax[0].imshow(mo_ret_0, cmap='gray')
ax[1].imshow(mo_ret_1, cmap='gray')
ax[2].imshow(cv_ret_0, cmap='gray')
ax[3].imshow(cv_ret_1, cmap='gray')
ax[4].imshow(mo_ret_0 - cv_ret_0, cmap='gray')
ax[0].set_title('sobelx in MONAI', fontsize=25)
ax[1].set_title('sobely in MONAI', fontsize=25)
ax[2].set_title('sobelx in CV2', fontsize=25)
ax[3].set_title('sobely in CV2', fontsize=25)
ax[4].set_title('diff mapx', fontsize=25)
If I change these two lines to without 1-
mo_ret_0 = 1 - (mo_ret[0] - np.min(mo_ret[0])) / (np.max(mo_ret[0]) - np.min(mo_ret[0]))
mo_ret_1 = 1 - (mo_ret[1] - np.min(mo_ret[1])) / (np.max(mo_ret[1]) - np.min(mo_ret[1]))
changed to
mo_ret_0 = (mo_ret[0] - np.min(mo_ret[0])) / (np.max(mo_ret[0]) - np.min(mo_ret[0]))
mo_ret_1 = (mo_ret[1] - np.min(mo_ret[1])) / (np.max(mo_ret[1]) - np.min(mo_ret[1]))
Results seem more reasonable.
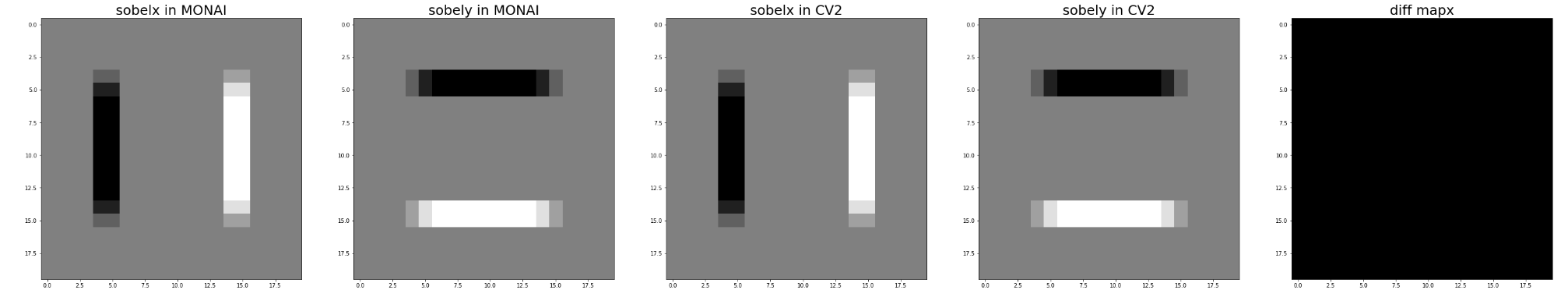

Thanks!
Hi @drbeh, I have just tested it with kernel_size=3 on fake data and data @JHancox shared with me. It seems we get the exact opposite of the value of OpenCV.

Here is my test code.
# fake data image = np.zeros([20,20]) image[5:15, 5:15] = 1 # real data image = hv_dir[0] filter = SobelGradients(kernel_size=3) mo_ret = filter(image[None]).squeeze() cv_ret_0 = cv2.Sobel(image, cv2.CV_64F, 1, 0, ksize=3) cv_ret_1 = cv2.Sobel(image, cv2.CV_64F, 0, 1, ksize=3) mo_ret_0 = 1 - (mo_ret[0] - np.min(mo_ret[0])) / (np.max(mo_ret[0]) - np.min(mo_ret[0])) mo_ret_1 = 1 - (mo_ret[1] - np.min(mo_ret[1])) / (np.max(mo_ret[1]) - np.min(mo_ret[1])) cv_ret_0 = 1 - (cv_ret_0 - np.min(cv_ret_0)) / (np.max(cv_ret_0) - np.min(cv_ret_0)) cv_ret_1 = 1 - (cv_ret_1 - np.min(cv_ret_1)) / (np.max(cv_ret_1) - np.min(cv_ret_1)) fig, ax = plt.subplots(1,5,figsize = (50,10)) ax[0].imshow(mo_ret_0, cmap='gray') ax[1].imshow(mo_ret_1, cmap='gray') ax[2].imshow(cv_ret_0, cmap='gray') ax[3].imshow(cv_ret_1, cmap='gray') ax[4].imshow(mo_ret_0 - cv_ret_0, cmap='gray') ax[0].set_title('sobelx in MONAI', fontsize=25) ax[1].set_title('sobely in MONAI', fontsize=25) ax[2].set_title('sobelx in CV2', fontsize=25) ax[3].set_title('sobely in CV2', fontsize=25) ax[4].set_title('diff mapx', fontsize=25)If I change these two lines to without
1-mo_ret_0 = 1 - (mo_ret[0] - np.min(mo_ret[0])) / (np.max(mo_ret[0]) - np.min(mo_ret[0])) mo_ret_1 = 1 - (mo_ret[1] - np.min(mo_ret[1])) / (np.max(mo_ret[1]) - np.min(mo_ret[1])) changed to mo_ret_0 = (mo_ret[0] - np.min(mo_ret[0])) / (np.max(mo_ret[0]) - np.min(mo_ret[0])) mo_ret_1 = (mo_ret[1] - np.min(mo_ret[1])) / (np.max(mo_ret[1]) - np.min(mo_ret[1]))Results seem more reasonable.

this looks like the difference between convolution and cross-correlation... perhaps the kernel should be flipped to make it consistent with the literature cc @kbressem who is working on https://github.com/Project-MONAI/MONAI/pull/5317
this looks like the difference between convolution and cross-correlation... perhaps the kernel should be flipped to make it consistent with the literature cc @kbressem who is working on #5317
When I originally ported this, I did find that using cross-correlation was the way to get the same result
@KumoLiu @wyli that's right, I flipped the differentiator kernel.
Actually what we have in deep learning as "convolution" is really "cross-correlation", which is similar except that the kernel is not flipped.
@wyli @Nic-Ma @KumoLiu any other comments here? Thanks
/black
/build
/build