Invalid json content on ds002735
Hello. Our dataset crawling service is detecting various OpenNeuro datasets containing invalid json files.
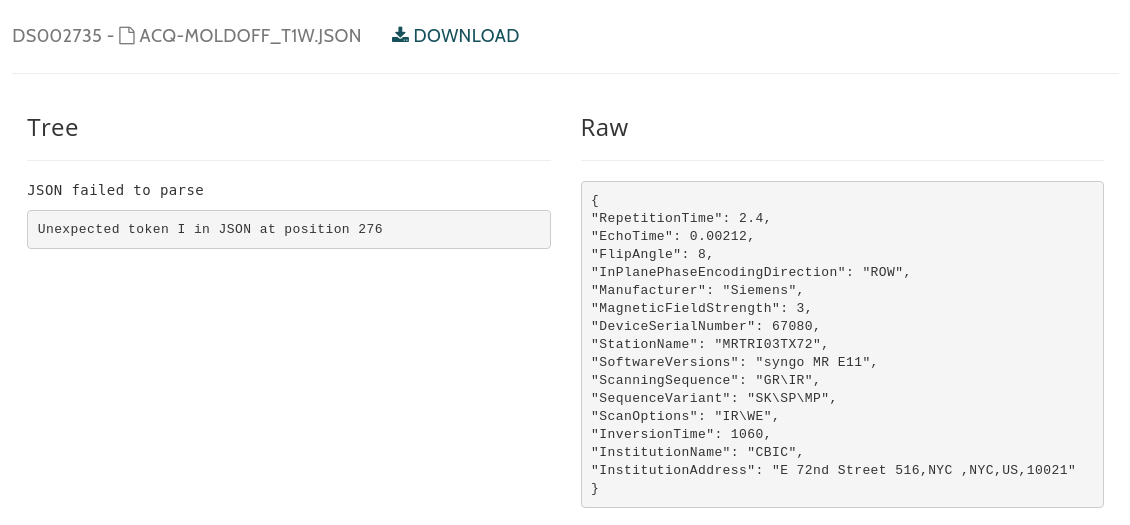
ailed to parse OpenNeuroDatasets/ds002735/acq-moldOFF_T1w.json
SyntaxError: Unexpected token I in JSON at position 276
failed to parse OpenNeuroDatasets/ds002041/task-rest_acq-fallypride_rec-acdyn_pet.json
SyntaxError: Unexpected token } in JSON at position 244
failed to parse OpenNeuroDatasets/ds000217/task-routelearning_events.json
SyntaxError: Unexpected token in JSON at position 354
failed to parse OpenNeuroDatasets/ds001241/._dataset_description.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse OpenNeuroDatasets/ds002336/channels.json
SyntaxError: Unexpected token } in JSON at position 314
failed to parse OpenNeuroDatasets/ds002549/._dataset_description.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse OpenNeuroDatasets/ds002718/._dataset_description.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse OpenNeuroDatasets/ds002338/channels.json
SyntaxError: Unexpected token } in JSON at position 314
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-009/._sub-009_scans.json
SyntaxError: Unexpected token in JSON at position 0
...
...
etc..
Right now I am ignoring those json files, but I wanted to report these issues in case someone might wants to know about them. I think the publisher of the data should correct these issues, or OpenNeuro to reject those .json files when submitted?
Maybe a BIDS validator issue? A place to start...
this should be caught by the validator - from OpenNeuro's perspective, we do not see these issues
I believe the validator works.. as I see the error message displayed here
https://openneuro.org/datasets/ds002735/versions/1.0.1/file-display/acq-moldOFF_T1w.json
Should OpenNeuro be publishing these files given they are known to be invalid? I think OpenNeuro should prevent user from submitting dataset with invalid JSON?
yep, though this should be the validators concern to identify these cases to ease remedying and head off downstream issues
FYI. I am still seeing some parse errors on some of the openneuro published datasets
(node:1091) DeprecationWarning: collection.ensureIndex is deprecated. Use createIndexes instead.
2020-08-04T04:07:37.297Z [undefined] [31merror [39m: read ECONNRESET
failed to parse OpenNeuroDatasets/ds002735/acq-moldOFF_T1w.json
SyntaxError: Unexpected token I in JSON at position 276
failed to parse OpenNeuroDatasets/ds002735/acq-moldON_T1w.json
SyntaxError: Unexpected token I in JSON at position 276
failed to parse OpenNeuroDatasets/ds002041/task-rest_acq-fallypride_rec-acdyn_pet.json
SyntaxError: Unexpected token } in JSON at position 244
failed to parse OpenNeuroDatasets/ds000217/task-routelearning_events.json
SyntaxError: Unexpected token in JSON at position 354
failed to parse OpenNeuroDatasets/ds001241/._dataset_description.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse OpenNeuroDatasets/ds002336/channels.json
SyntaxError: Unexpected token } in JSON at position 314
failed to parse OpenNeuroDatasets/ds002549/._dataset_description.json
SyntaxError: Unexpected token in JSON at position 0
2phasemag given with only phase1?
failed to parse OpenNeuroDatasets/ds002718/._dataset_description.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse OpenNeuroDatasets/ds002338/channels.json
SyntaxError: Unexpected token } in JSON at position 314
description.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse OpenNeuroDatasets/ds001840/task-viewclips_eyetrack.json
SyntaxError: Unexpected token } in JSON at position 177
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-001/._sub-001_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-002/._sub-002_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-003/._sub-003_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-004/._sub-004_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-005/._sub-005_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-006/._sub-006_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-007/._sub-007_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-008/._sub-008_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-009/._sub-009_scans.json
SyntaxError: Unexpected token in JSON at position 0
Should there be a mechanism for OpenNeuro to remove files that are invalid (files with issues that didn't get detected when they were uploaded?)
I think the approach of adding a notification to the validator is reasonable, perhaps @rwblair may have some thoughts
This should be solved in the validator.
How should I handle invalid JSON files that are already published in the OpenNeuro? Is there a plan to purge them from the OpenNeuro, or should I come up with "ignore" file that list those invalid JSON?
failed to parse OpenNeuroDatasets/ds002735/acq-moldOFF_T1w.json
SyntaxError: Unexpected token I in JSON at position 276
failed to parse OpenNeuroDatasets/ds002735/acq-moldON_T1w.json
SyntaxError: Unexpected token I in JSON at position 276
failed to parse OpenNeuroDatasets/ds002041/task-rest_acq-fallypride_rec-acdyn_pet.json
SyntaxError: Unexpected token } in JSON at position 244
failed to parse OpenNeuroDatasets/ds000217/task-routelearning_events.json
SyntaxError: Unexpected token in JSON at position 354
failed to parse OpenNeuroDatasets/ds001241/._dataset_description.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse OpenNeuroDatasets/ds002336/channels.json
SyntaxError: Unexpected token } in JSON at position 314
failed to parse OpenNeuroDatasets/ds002549/._dataset_description.json
SyntaxError: Unexpected token in JSON at position 0
2phasemag given with only phase1?
failed to parse OpenNeuroDatasets/ds002718/._dataset_description.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse OpenNeuroDatasets/ds002338/channels.json
SyntaxError: Unexpected token } in JSON at position 314
failed to parse OpenNeuroDatasets/ds001997/._dataset_description.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse OpenNeuroDatasets/ds001840/task-viewclips_eyetrack.json
SyntaxError: Unexpected token } in JSON at position 177
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-001/._sub-001_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-002/._sub-002_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-003/._sub-003_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-004/._sub-004_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-005/._sub-005_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-006/._sub-006_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-007/._sub-007_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-008/._sub-008_scans.json
SyntaxError: Unexpected token in JSON at position 0
failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-009/._sub-009_scans.json
SyntaxError: Unexpected token in JSON at position 0
If it's this few, perhaps we should just fix them ourselves...
Part of the problem is that the validator has a built in implicit ignore list that includes all dot files, among other things. The is also an issue with how errors for invalid syntax in specifically sidecar json files are reported in the validator, will create an issue and try and fix this.
Should the validator try to read in any and all json files regardless of bidsignore?
Should the uploader only upload the same list of files that the validator tests?
Should the uploader only upload the same list of files that the validator tests?
We should not be uploading implicitly ignored files. I think #1656/#1797 should be addressing that, but we should probably remove the ones that slipped through.
@soichih by chance is this the full list of problematic files you all have found? until we have the solution deployed I can take a pass at remedying this
@franklin-feingold Yes, it looks like those are the only problematic files that we are finding.
@soichih I went through and resolved your list (tracked below) by making patch releases. I commented on a few datasets.
-
[x] failed to parse OpenNeuroDatasets/ds002735/acq-moldOFF_T1w.json SyntaxError: Unexpected token I in JSON at position 276
-
[x] failed to parse OpenNeuroDatasets/ds002735/acq-moldON_T1w.json SyntaxError: Unexpected token I in JSON at position 276
-
[x] failed to parse OpenNeuroDatasets/ds002041/task-rest_acq-fallypride_rec-acdyn_pet.json SyntaxError: Unexpected token } in JSON at position 244
-
[x] failed to parse OpenNeuroDatasets/ds000217/task-routelearning_events.json SyntaxError: Unexpected token in JSON at position 354
-
[x] failed to parse OpenNeuroDatasets/ds001241/._dataset_description.json SyntaxError: Unexpected token in JSON at position 0
-
[x] failed to parse OpenNeuroDatasets/ds002336/channels.json SyntaxError: Unexpected token } in JSON at position 314
-
[x] failed to parse OpenNeuroDatasets/ds002549/._dataset_description.json SyntaxError: Unexpected token in JSON at position 0 2phasemag given with only phase1?
-
[ ] failed to parse OpenNeuroDatasets/ds002718/._dataset_description.json SyntaxError: Unexpected token in JSON at position 0 FF: This file appears to be removed in the latest version of the dataset
-
[x] failed to parse OpenNeuroDatasets/ds002338/channels.json SyntaxError: Unexpected token } in JSON at position 314
-
[ ] failed to parse OpenNeuroDatasets/ds001997/._dataset_description.json SyntaxError: Unexpected token in JSON at position 0 FF: This dataset was removed
-
[x] failed to parse OpenNeuroDatasets/ds001840/task-viewclips_eyetrack.json SyntaxError: Unexpected token } in JSON at position 177
FF: ds002507 was removed
-
[ ] failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-001/._sub-001_scans.json SyntaxError: Unexpected token in JSON at position 0
-
[ ] failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-002/._sub-002_scans.json SyntaxError: Unexpected token in JSON at position 0
-
[ ] failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-003/._sub-003_scans.json SyntaxError: Unexpected token in JSON at position 0
-
[ ] failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-004/._sub-004_scans.json SyntaxError: Unexpected token in JSON at position 0
-
[ ] failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-005/._sub-005_scans.json SyntaxError: Unexpected token in JSON at position 0
-
[ ] failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-006/._sub-006_scans.json SyntaxError: Unexpected token in JSON at position 0
-
[ ] failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-007/._sub-007_scans.json SyntaxError: Unexpected token in JSON at position 0
-
[ ] failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-008/._sub-008_scans.json SyntaxError: Unexpected token in JSON at position 0
-
[ ] failed to parse subject level json: OpenNeuroDatasets/ds002507/sub-009/._sub-009_scans.json SyntaxError: Unexpected token in JSON at position 0
Here's the results of my current test (jq null < $JSON):
ds000224/sourcedata/phenotype/BAS_BIS.json
parse error: Invalid numeric literal at line 6, column 20
ds000224/sourcedata/phenotype/KBIT2.json
parse error: Invalid numeric literal at line 6, column 32
ds000224/sourcedata/phenotype/NEO_
zsh: no such file or directory: ds000224/sourcedata/phenotype/NEO_
ds000224/sourcedata/phenotype/NIH_Toolbox.json
parse error: Invalid numeric literal at line 6, column 54
ds001415/sourcedata/behavioral/task-maplistening_psychopy.json
parse error: Expected separator between values at line 105, column 17
ds001785/sub-patient/ses-01/ieeg/sub-patient_ses-01_space-postimplant_coordsystem.json
parse error: Expected another key-value pair at line 9, column 1
ds002330/acq-lr_dwi.json
parse error: Invalid escape at line 22, column 27
ds002330/acq-rl_dwi.json
parse error: Invalid escape at line 22, column 27
ds002330/dir-ap_epi.json
parse error: Invalid escape at line 23, column 24
ds002330/dir-pa_epi.json
parse error: Invalid escape at line 23, column 24
ds002330/T1w.json
parse error: Invalid escape at line 20, column 28
ds002330/T2w.json
parse error: Invalid escape at line 22, column 31
ds003090/derivatives/manual-masks/dataset_description.json
parse error: Expected another key-value pair at line 9, column 5
ds003242/scans.json
parse error: Expected separator between values at line 7, column 21
ds003459/task-visortho_bold.json
parse error: Invalid escape at line 4, column 2080
ds003459/task-visphono_bold.json
parse error: Invalid escape at line 4, column 2024
ds003459/task-vissem_bold.json
parse error: Invalid escape at line 4, column 1628
ds003459/task-vissynt_bold.json
parse error: Invalid escape at line 4, column 1591
ds003481/project_descriptions/Speech-act_Emotion_categorization.json
parse error: Expected separator between values at line 4, column 1239
ds003481/project_descriptions/Speech-act_recognition.json
parse error: Expected separator between values at line 5, column 1165