echopype
 echopype copied to clipboard
echopype copied to clipboard
Converting large data files
During OceanHackWeek'21 a participant attempted to convert ~1GB files from Nortek AD2CP (Signature 100 and Signature 250), and failed both on the Jupyter Hub and on their personal machine. This is probably related to the exploding in-memory xarray.merge situation that we have seen.
@imranmaj @lsetiawan and I discussed this yesterday, and an interim solution is to have an under the hood procedure to do the following:
- parse binary data
- if parsed binary data reaches a certain size, save already parsed data into a small file
- repeat 1-2 until end of binary file
- merge all small files under the hood, which can be delayed and distributed to dask workers.
Step 4 likely will use the same mechanisms as the combine EchoData method that is currently in an PR (#383), though it will use individual smaller EchoData objects obtained via open_converted rather than via open_raw (the difference is whether the groups are lazy-loaded or persist in memory)
@imranmaj will take a stab at this for AD2CP files, but I think the results can serve as a good road map for other sonar models, since the only difference really is how the binary file is parsed. @imranmaj : it'll be great if you could think about this parallel structure when you develop the solution for AD2CP!
Wanted to document our discussions so far, as the starting point of conversation as we find a good solution.
Bringing back attention to this issue: we have similar needs for EK files, probably not quite on local machine unless people have very large files, but on environment with smaller RAM this is an issue with kinda-large files.
Testing with a 725 MB file from OOI:
- on my machine the highest memory load was about 4.5 GB and in multiple waves
- on aws pangeo hub: kernel dies when doing open_raw
- on binder: kernel dies when doing open_raw
- on pangeo binder: kernel dies when doing open_raw
Tagging @lsetiawan per our discussion today.
I did something like this last year for a different project, though not in a parallelized fashion. Here's a link to the code (a notebook), in case there's something of interest there. It reads data in sequential chunks from a large, local binary file (23 GB), and sequentially appends to a Zarr dataset on either AWS or locally, after creating the zarr dataset on the first chunk. Thanks go to @lsetiawan for help and actual code, plus to a blog post from Rich Signell that I can't quite locate anymore.
The local module that's imported is https://github.com/snowmodel-tools/postprocess_python/blob/master/awszarr/snowmodelzarrfs.py. That just handles file I/O for AWS vs local.
Hello, I am working for NOAA NCEI with the goal of processing 100+ TB of EK60 data (currently found in the AWS Open Data Registry).
We would like to do all of our processing with lambda functions, converting raw EK60 files to Zarr in the cloud.
Currently we are having the same issues mentioned above with memory requirements exceeding available resources.
Using a profiler, I have analyzed echopype opening and processing "L0003-D20040909-T161906-EK60.raw," a 95 MB file known to crash an AWS lambda using full resources allocated (10,240 MB memory and 900 second timeout):
Line-by-line memory usage:
$ python -m memory_profiler process_sv_data.py
Line # Mem usage Increment Occurences Line Contents
============================================================
4 102.531 MiB 102.531 MiB 1 @profile
5 def process_data():
6 102.531 MiB 0.000 MiB 1 raw_file_s3path = "s3://noaa-wcsd-pds/data/raw/Albatross_Iv/AL0409/EK60/L0003-D20040909-T161906-EK60.raw"
7 3105.227 MiB 3002.695 MiB 1 echodata = ep.open_raw(raw_file_s3path, sonar_model='EK60', storage_options={'anon': True})
8 2940.949 MiB -164.277 MiB 1 ds_Sv = ep.calibrate.compute_Sv(echodata)
9 2941.074 MiB 0.125 MiB 1 print(ds_Sv.Sv.shape)
10 5348.723 MiB 2407.648 MiB 1 ds_Sv.to_zarr(store="L0003-D20040909-T161906-EK60.zarr")
11 5348.723 MiB 0.000 MiB 1 return
The echodata and ds_Sv objects are each respectively 3 and 2 GB in size.
Another profile below shows dynamic memory usage when (1) opening the file, (2) calibrating it, and (3) writing to zarr:
$ mprof run process_data.py
$ mprof plot
import echopype as ep
@profile
def read_from_bucket():
raw_file_s3path = "s3://noaa-wcsd-pds/data/raw/Albatross_Iv/AL0409/EK60/L0003-D20040909-T161906-EK60.raw"
return ep.open_raw(raw_file_s3path, sonar_model='EK60', storage_options={'anon': True})
@profile
def calibrate_data(echodata):
return ep.calibrate.compute_Sv(echodata)
@profile
def save_to_zarr(ds_Sv):
ds_Sv.to_zarr(store="L0003-D20040909-T161906-EK60.zarr")
if __name__ == '__main__':
echodata = read_from_bucket()
ds_Sv = calibrate_data(echodata)
save_to_zarr(ds_Sv)
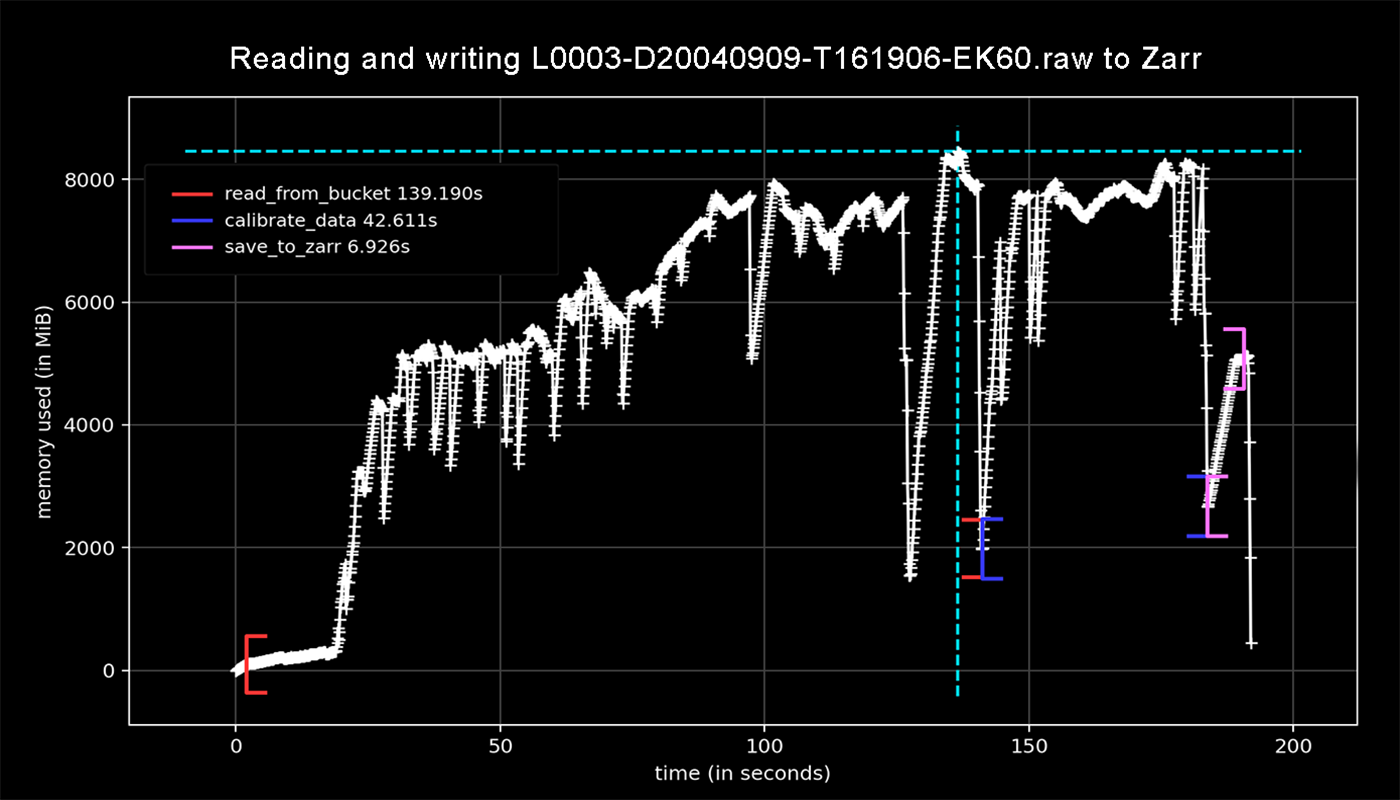
The plot shows the 95 MB file is using ~9 GB of memory to read the raw data and similar to calibrate it! With memory polling only every 0.1 seconds I would presume it spikes even above this, crashing the lambda.
Our team would like to pursue @leewujung's interim solution, writing out smaller files while processing the raw data. With many of the EK60 files in the dataset listed above measuring 500 MB or more in size we are going to need a feature to accommodate their processing without needing high-memory resources (3 GB is currently the largest raw file in the bucket).
I would like to help work on this problem so we can expedite implementing a solution but I am new to your code-base. I am thinking a starting point would be to modify the method reading the datagrams to limit it to subsets of the data. It would be very helpful to me if I could get some guidance on the best way to approach this so that the code will be useful to the community.
@oftfrfbf : Great that you are looking into this! Welcome onboard!
I think there are actually 2 separate issues here:
-
A truly large raw data file, like a 9.5 GB (!) AD2CP file, or the 3 GB EK60 file you mentioned: This likely would require the approach outlined in the first comment above
-
An actually not very big file, like the 95 MB EK60 file you mentioned: We should do a profile under the hood to see whether it is the raw file parsing component or the part where data are organized into xarray datasets that is expanding the memory (I suspect it is the latter). If it is the latter, we have been discussing experimenting with the dask delayed functionality to see if the operations can be distributed to different workers
Could you try doing a memory profile under the hood of open_raw? (for 2. above)
The functions being called are here:
https://github.com/OSOceanAcoustics/echopype/blob/main/echopype/convert/api.py#L412-L445
parser deals with the actual parsing of the data files, and the setgrouper deals with assembling the datasets.
Thanks, @leewujung I will work on some further profiling and post results here.
Had to shift some things around with closures so I could use the profile decorator but here are some results.
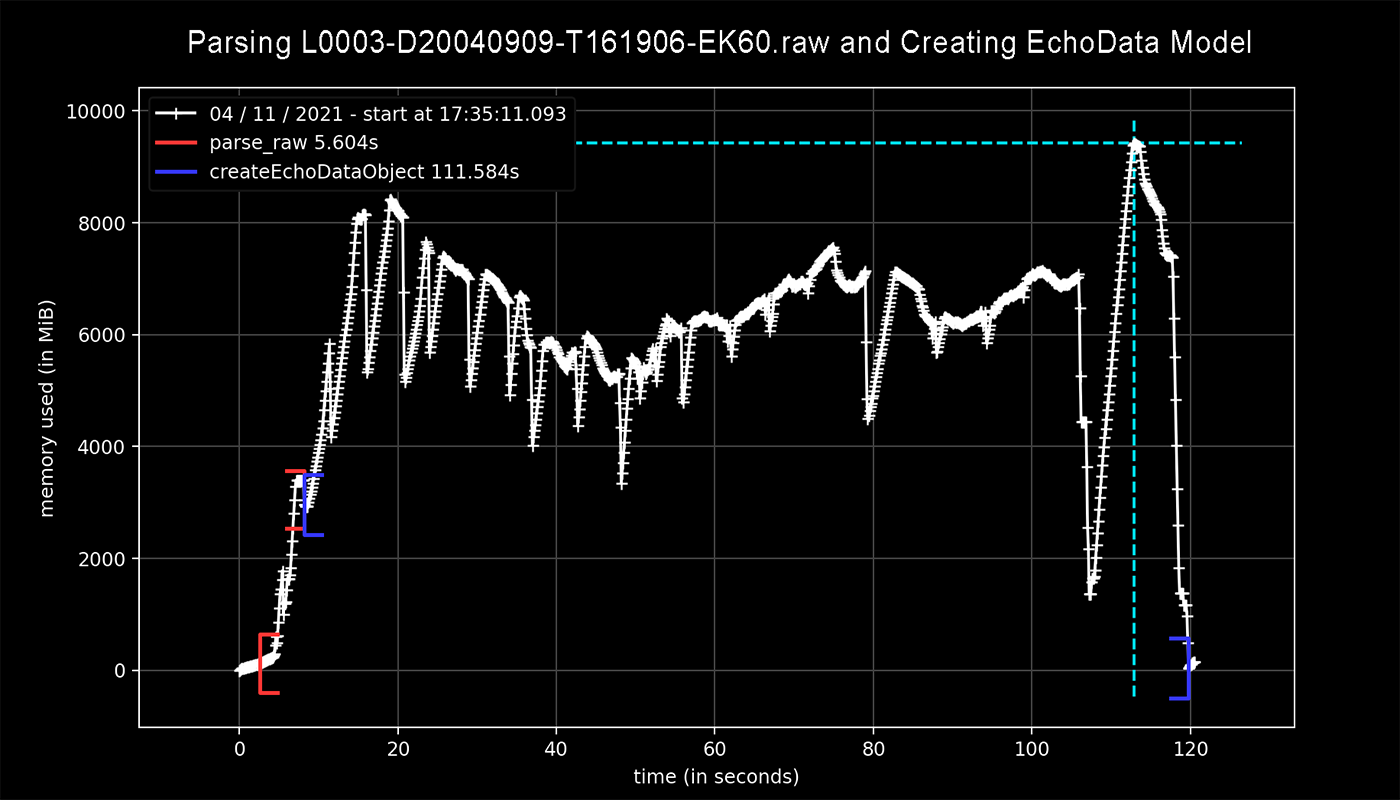
Profiling for parser.parse_raw() (shown in red) only uses about 3 GB of memory while lines 424 through 445, accounting for the creation of the EchoData model (shown in blue), brought that value up to a total of 10 GB.
The memory expansion issue (from 2. above) is now split to a new issue #489 .
The latest update on addressing the memory expansion is in dev now via #1185 and this example file has been used as a test data file. I'll close this now as files from Simrad echosounders are now taken care of, and open a new one for AD2CP files, which was what this issue was originally for.