Viewers
 Viewers copied to clipboard
Viewers copied to clipboard
Can't load multiframe XA
Bug Report
Version: v3, v2
Describe the Bug
Impossible to use the viewer with XA (Coro) studies.
What steps can we follow to reproduce the bug?
- https://ohif.org/get-started
- Select 'Use your own data'
- Upload the enclosed sample (i reproduce with all my files :'() https://filetransfer.io/data-package/DAye0RDw#link
- Use the cine play button or scroll: the image count change, the image displayed stay the first one.
The sample image has no pixel data (i.e., is missing element 7fe0,0010). This is required for the SOP Class UID in the meta-data (X-Ray Angiographic Image Storage)
It does have an IconImageSequence (element 0088,0200) that shows it was probably created by Applicare/RadWorks/Version 5.0 and the pixel data element is buried in that sequence.
My guess this is not so much an OHIF Viewer issue but a non-standard DICOM image issue from your source. It does not display on some other DICOM viewers either (e.g., Horos) while some do seem to handle this non-standard image (e.g., Weasis).
Todd Jensen, PhD Jensen Informatics LLC
Hello Todd, indeed, it should be images from GE CA1000, not a very recent device, but you know how long it can be for hospitals to move to more recent systems. Other viewers indeed appear to have no issues with it (playing with MedDream for exemple, perfect handling of it).
Do you believe that it's a complete no-go for ohif to implement the support of it? If so, ohif can't be used reliably, at least in cardiology and radiology in some countries (in France this GE system is used in the majority of hospitals and clinics)
Hello @jensen0914 ,
I try to figure out my options here and i have a difficulty:
The sample image has no pixel data (i.e., is missing element 7fe0,0010).
By looking at it with dcmtk editor i can see the 07fe0,0010 tag, with the pixel data inside it and it can be exported with success.
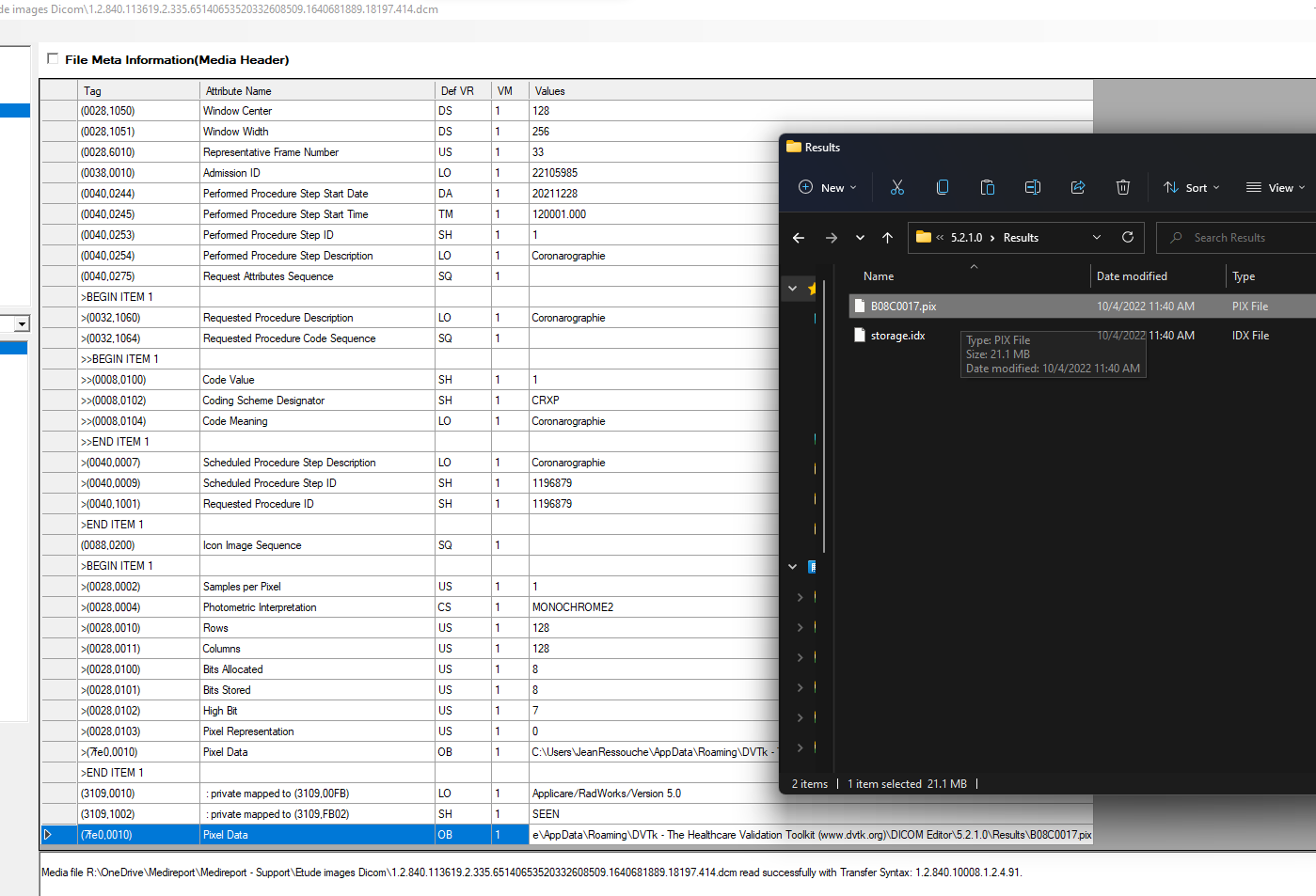
Do you believe that DCMTK is doing some internal magic to recover automatically the pixel data from the IconImageSequence ?
According to the doc, 0088,0200 is used for the thumbnail.
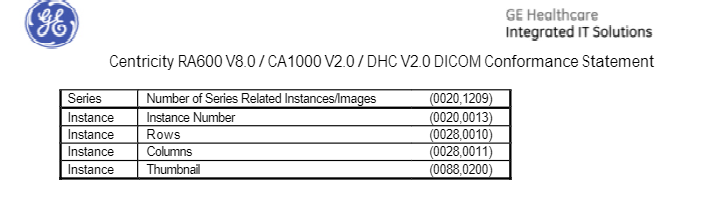
I was mistaken, the file does have pixel data - I'm not sure why my first run missed seeing that.
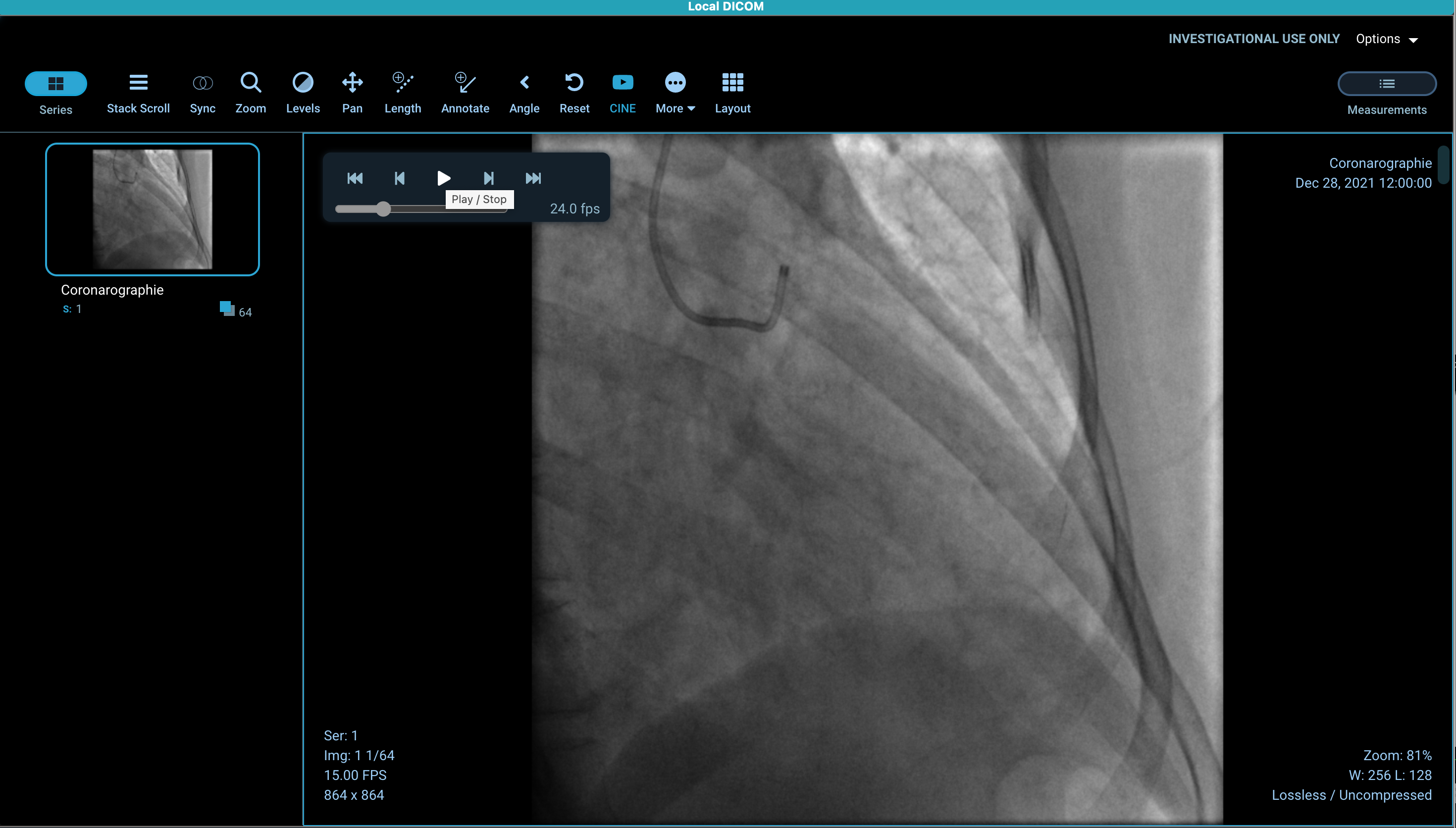
BTW - I can view this file when I load it in OHIF Viewer I build from this forked branch (jensen0914/Viewers](https://github.com/jensen0914/Viewers/tree/saincepacs-ohif-viewer), which is basically only a few commits behind OHIF Viewer v2 master branch (4.12.37 vs 4.12.41).
And in the v3 get started link:

Hi @jensen0914 , thanks again for your input. i can also see it but i only see 1 frame. With this version you can play the entire sequence? On my side playing or scrolling to display the next frame is not working.
In the v3 demo, I can only see the first frame even when scrolling. In my v2 build, I can see all of the frames when scrolling or using cine:
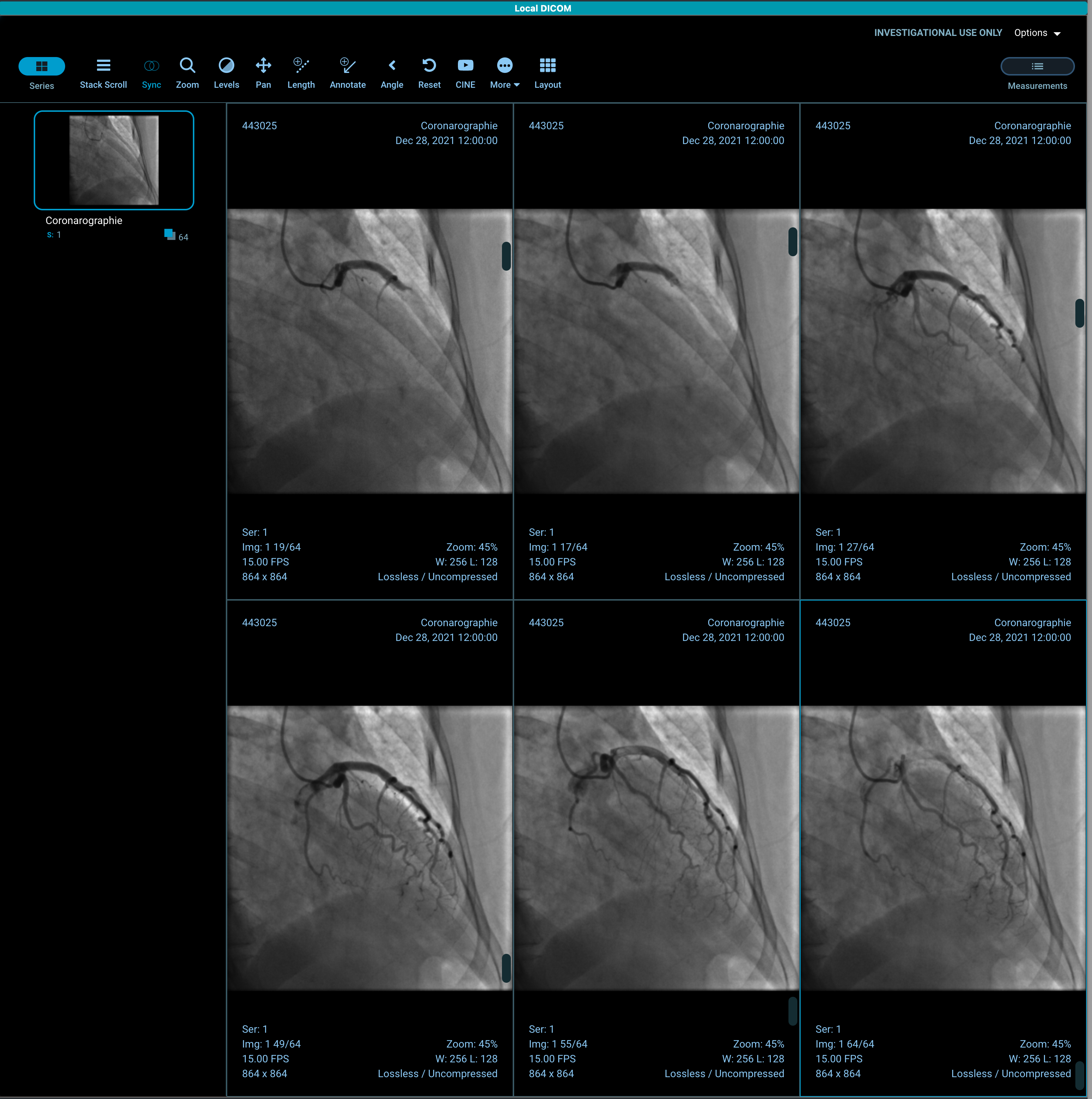
Are you loading the data using JSON?
Thanks @jensen0914, then i'll try with the v2 again, building it myself as you did. @arturojain nope, loading from a local .dcm file on the v3 demo version and from a dcm4chee source on my environment, with the same result.
Could you kindly provide the data if it has been anonymized and you can confirm that there is no patient health information present in any of the headers or embedded within the pixel data?
can you re-upload for us please
Hello, I'm facing the same issue. I can't seem to run the Angiographic images (XA modalities). Has anyone found a solution or determined the cause of this problem?
Thanks!