[Feature] Merging "Total spiking probability edges" into elephant
Feature
Hello, I would like to merge a new algorithm called Total spiking probability edges into elephant.
TSPE enables the classification of excitatory and inhibitory synaptic effects using a connectivity estimation. It uses the cross-correlation of spiketrains at different delays combined with specially designed Edge-Filters to distinguish between excitatory and inhibitory connections.
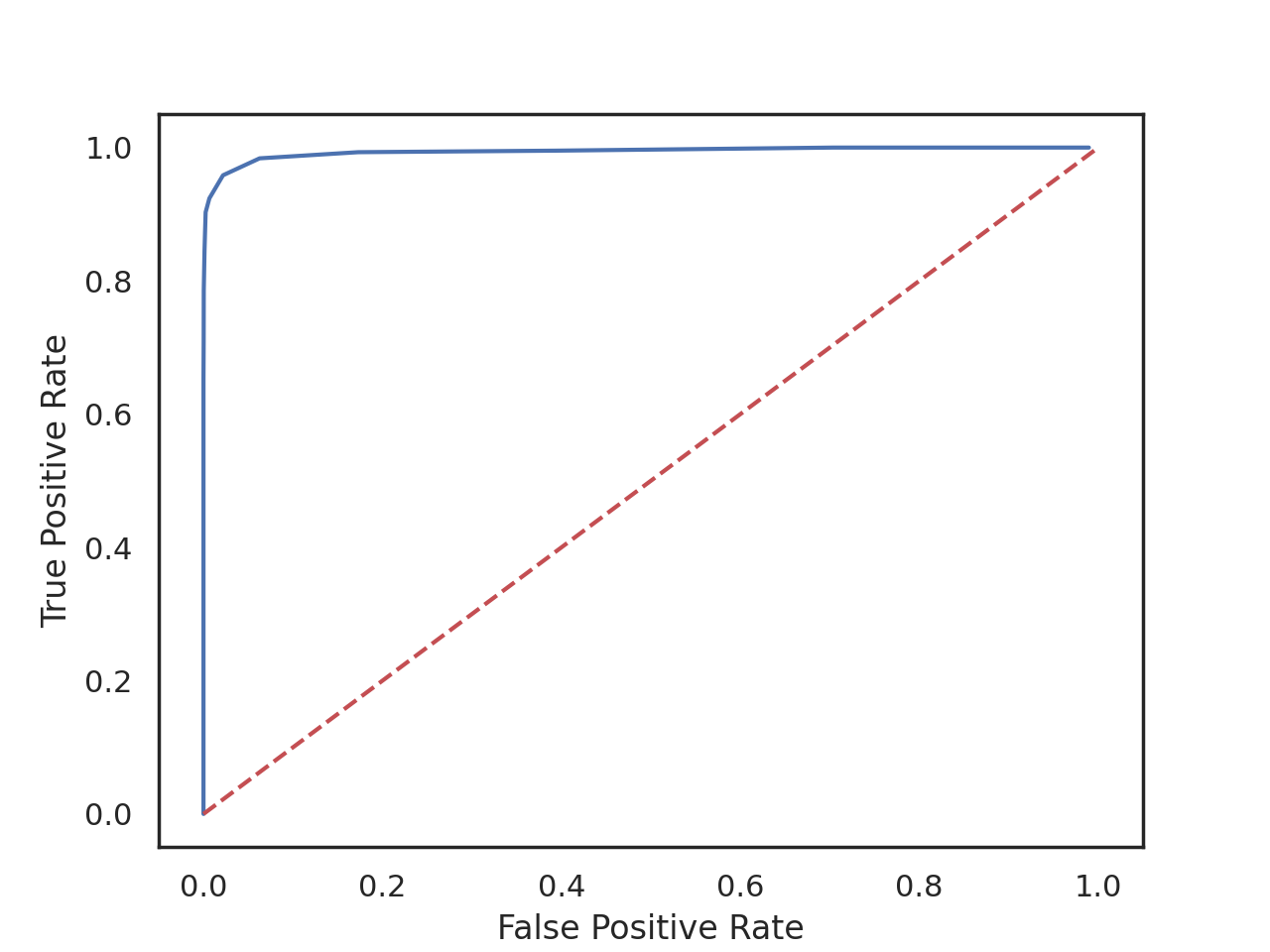
TSPE was evaluated with large scale in silico networks and enables almost perfect reconstructions (true positive rate of approx. 99% at a false positive rate of 1% for low density random networks) depending on the network topology and the spike train duration.
Motivation This project was originally implemented using MATLAB (https://github.com/biomemsLAB/TSPE). To open up the project into the Python-ecosystem, I was tasked to create a reference-implementation of this algorithm in Python as part of my student research paper. Since the laboratory where I'm doing this uses the elephant-project, we thought about including it directly in the project itself.
Additional context The algorithm is already fully implemented by me with the exception of the normalization across all SPE (see SPE' in the paper) and proper tests using pytest. The algorithm performs as well as the original implementation and when using the evaluation used by the original paper it yields similar results. The code can be found here: https://github.com/zottelsheep/tspe-python
If there's interest into including this into elephant, I would be happy to implement further tests to comply with the contribution-guidelines, but I'd still need some guidance as to where to put the code, interface wishes, etc.
The only problem is that I need to finish this by the beginning of May, since I have to finish my paper by then.
Here are some graphs from TSPE evaluated using the ExampleSpikeTrain given in the original implementation:

Hi @zottelsheep , first of all, thank you so much for your input.
Please excuse the delayed response due to Easter vacation and a number of business trips.
We were not aware of this method, but it certainly looks very interesting and definitely falls into the scope of Elephant. At first glance, it seems to us that the function would likely open up a new module (perhaps functional_connectivity_estimation?) in the Elephant library. Also, a few things regarding inputs would need to be adapted. Please allow us some days to look into your code and make concrete suggestions how to proceed with opening a pull request.
As we are currently still on travel, we were not able to obtain the reference manuscript as it is currently not in our online institutional subscription. Is there a preprint available by chance?
Thanks for the response!
Yes, please have a look at the code. I know its quite soon, but do you think it would be possible to give me some feedback by next Tuesday? Mainly to get an idea how I need to plan my roadmap for the rest of my paper.
Regarding the original paper: I did not find a preprint but the original master-thesis is available here https://arxiv.org/abs/2005.07083. Section four and five are basically the publication I have linked above.
Hello @zottelsheep,,
Thank you for providing the master thesis as a reference.
As mentioned earlier the roadmap could include new module called functional_connectivity consisting of the file elephant/functional_connectivity.py used only for importing function calls, and a folder elephant/functional_connectivity_src/total_spiking_probability_edges.py which contains the main function total_spiking_probability_edges.
Unit tests could be contained in a corresponding test module called test_total_spiking_probability_edges.py in elephant/test. It would be great if you could open a pull request to add those modules to Elephant.
Refer to this very crude sketch of a possible implementation of functional_connectivity.py:
from elephant.functional_connectivity_src.total_spiking_probability_edges import total_spiking_probability_edges
__all__ = [
"total_spiking_probability_edges"
]
[...]
and elephant/functional_connectivity_src/total_spiking_probability_edges.py:
def total_spiking_probability_edges(spike_trains,
a: Optional[List[int]] = None,
b: Optional[List[int]] = None,
c: Optional[List[int]] = None,
max_delay: int = 25,
[..])
"""
Performs the Total spiking probability edges (TSPE) :cite:`tspe-de_blasi2007_???` on spiketrains ...
Parameters
----------
spiketrains: neo.spiketrains
Returns
-------
tspe_matrix
"""
# implementation pseudo-code
normalized_cross_correlation=normalized_cross_correlation(spiketrains, [...])
[..]
tspe_matrix = filter_edges(normalized_cross_correlation)
return tspe_matrix
def normalized_cross_correlation() [..]
def filter_edges() [...]
In a later stage, we would be happy to check if normalized_cross_correlation could be replaced or merged with Elephant's cross correlation function.
Instead of parameters a, b and c, it would be nice to have more telling names -- would you have a suggestion? Maybe something like filter_edge_a or 'filter_segment_a' or something like that?
Also, your reference can be added to the Elephant bibliography file, located at elephant/doc/bib/. For instructions on how to add this, see this guide: https://elephant.readthedocs.io/en/latest/contribute.html
In case you need assistance in writing the unit tests, we would be more than happy to help. However, if you are able to supply some unit tests, that would be even better.
The examples provided under scripts/test_tspe.py would make for a nice validation test. Testing the python implementation against some ground truth or against the original matlab implementation. The ground truth data or the results calculated with matlab could be uploaded to the elephant-data repository and a unit test could be written comparing the calculated results using the new implementation against the data from elephant data, see here https://gin.g-node.org/NeuralEnsemble/elephant-data .
Once again, thank you for your contribution and we look forward to hearing from you soon.
In Summary: Create a pull request as a starting point with the aforementioned modules, and we can work our way including adding docstrings and unit tests from there.
Note: Please be advised that your code will be published under the BSD-3-clause license which is more permissive than your previously used GNU license.
Hey :)
That sounds great and quite doable. Thanks for the pointers, I'll open up a pull-request soon.
As for the filter_names. I agree that we could name them more clearly. I stuck to a, b, c as that is what was used in the paper. The matlab-implementation used the following names but I think they're a bit confusing:
neg_wins - Windows for before and after area of interest (default [3, 4, 5, 6, 7, 8])
co_wins - Cross-over window size (default 0)
pos_wins - Sizes of area of interest (default [2, 3, 4, 5, 6])
I've been thinking about something along the lines of observed_window_size, crossover_window_size and surrounding_window_size, but I'm not sure if we should leave the a,b and c somewhere in there? But we can discuss this further in the actual pull-request^^
Since I've last written you, I have also included a test in my repo for testing tspe against some sample-data provided in the original repo. It basically checks if the auc is greater than 95%.
And thanks for the info about the license. While I don't have a problem with the license change on my code, I will need to check how we can use the validation-dataset since I don't own the rights for it. Or is this license managed separately?
Hi, cool, we're looking forward to the PR.
Regarding the license of the validation dataset, this can be handled more flexible. Typically, we would suggest to put data under a CC-BY 4.0 license, and any accompanying code (e.g., a piece of code producing the desired result using matlab; or a piece of code generating the validation data, ...) under BSD 3-clause for simplicity -- but that could also be handled differently (we put a license on a per-directory basis there).