Integrating AdvancedHMC
I'd like to give a try on integrating Poriot with AdvancedHMC when I find some time. Can you give me some brief intro of how it should be done?
using Poirot, IRTools
coin() = rand(Bool)
f = () -> begin
a = coin()
b = coin()
observe(a | b)
a & b
end
Get a trace / static graph for the generative model:
julia> ir = Poirot.trace(typeof(f))
1: (%1 :: var"#9#10")
%2 = (rand)(Bernoulli{Rational{Int64}}(p=1//2)) :: Bool
%3 = (rand)(Bernoulli{Rational{Int64}}(p=1//2)) :: Bool
%4 = (|)(%2, %3) :: Bool
%5 = (Poirot.observe)(%4) :: Nothing
%6 = (&)(%2, %3) :: Bool
return %6
Transform this into a logpdf, along with a list of random variables and their types:
julia> lpdf, vars = Poirot.logprob(ir);
julia> vars
2-element Array{Any,1}:
(%2, Bool)
(%3, Bool)
Transform this into an executable Julia function and run it (it returns both the result of the infer block and a log probability for that result):
julia> lpdf = IRTools.func(lpdf)
##254 (generic function with 1 method)
julia> lpdf(nothing, (true, false))
(false, -1.3862943611198906)
julia> lpdf(nothing, (false, false))
(false, -Inf)
You should be able to treat lpdf as a normal function, including differentiating with Zygote to get a gradient. I believe that's all you need for HMC but let me know if there's something else.
Somehow came across this old issue and play with Poirot a bit today. I managed to make a model with discrete observations work with HMC---see below.
sigmoid(x) = 1 / (1 + exp(-x))
f = () -> begin
l = randn()
p = sigmoid(l)
a = rand(Bernoulli(p))
b = rand(Bernoulli(p))
c = rand(Bernoulli(p))
end
ir = Poirot.trace(typeof(f))
_lpdf, vars = Poirot.logprob(ir)
_lpdf = IRTools.func(_lpdf)
lpdf = x -> _lpdf(nothing, (x[1], true, true, false))[2] # observations are a=true, b=true, c=false
gpdf = x -> only(Zygote.gradient(lpdf, x))
using AdvancedHMC, ForwardDiff
dim = 1
xinit = randn(dim)
n_samples = 2_000
metric = DiagEuclideanMetric(dim)
hamiltonian = Hamiltonian(metric, lpdf, ForwardDiff)
integrator = Leapfrog(0.5)
proposal = NUTS{MultinomialTS, GeneralisedNoUTurn}(integrator)
samples, stats = sample(hamiltonian, proposal, xinit, n_samples; verbose=false)
The sample histogram looks correct to me:
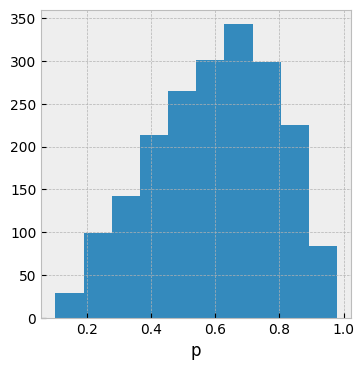
In the meanwhile, I don't know how to make a simple model with continuous observation work, e.g. below fails by not moving at all in the sampling
f = () -> begin
m = randn()
x = rand(Normal(m, 1))
end
# ...
lpdf = x -> _lpdf(nothing, (x[1], 1.0))[2]
but I'd expect to sample m around 1.0.