Support nice interpolation for C_ells
Related to #641 if one tries to compute power spectra at all ells, the computation becomes super slow. In practice one never needs to do this. You evaluate the power spectrum at a few ells and then interpolate.
It's been proposed that we create a framework to manage these interpolations so users don't have to create their own.
Well in Angpow I had a mechanism to generate ell list like
ell=0/1/2/3/4/5/6/7/8/9/10/11/12/13/14/16/18/20/22/25/28/32/36/41/47/54/62/71/81/93/106/121/139/159/182/209/240/275/315/355/395/435/475/515/555/595/635/675/715/755/795/835/875/915/955/995/999/
with a linear part at low ell and a logscale a high ell. Then, from the list of (ell_i, Cl_i) I use a spline interpolation to get (ell_j, Cl_j) with all ell_j from 0 to 999.
Now. to implemnt in CCL if no bodyelse take care of this issue, I would like to know:
- do you expect a C-code or a Python one for pyccl
- the generation of (ell_i, Cl_i) list first needs 2 parameters to produce the ell_i list, then the cl computation needs to use such a list, and finally here one addresses only 1 cl computation but I guess you need Cl for each couple of tracers, each couple of bins, so there is may be in CCL a mechaninsm to get the collection of Cls produced, but then each Cl^{ab}(z1,z2) list should be associated to the ell_i list. Does this exists already?
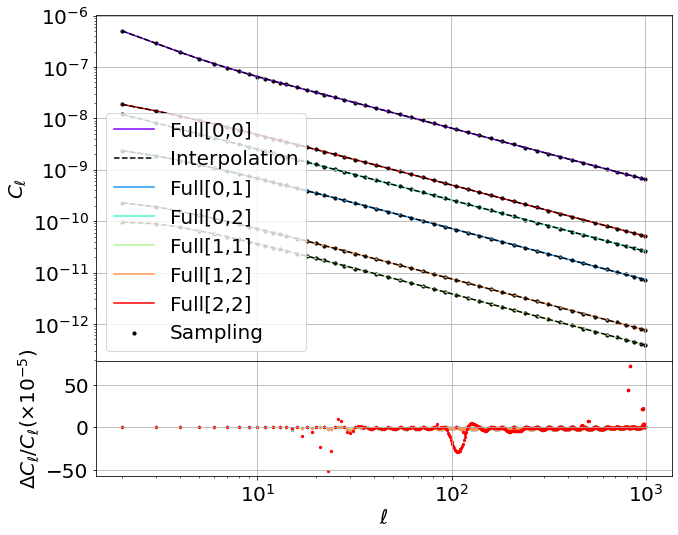
I have cooked a Google Collab nb to show what can be done: have a look here
With ~ 50 ells sampling values in the range [2, 1000] for different tracers Cell auto-cross.