sharing strange smudgeplot for imporve smudgeplot
hi, everyone
i running smudgeplot for my data, get strange smudgeplot. look the below
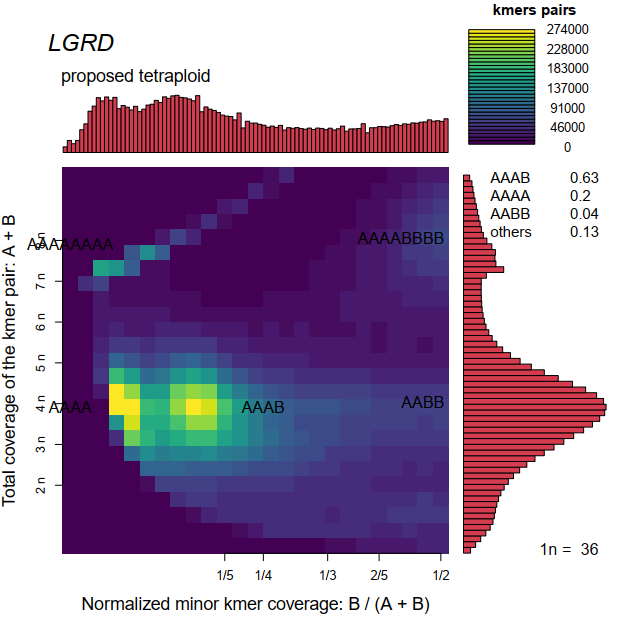
i read the smudgeplot_warnings.txt, get information about provide the information for imporve the software. so i open this is issue. How to interpret this result?
Hi,
thanks for posting. This genome indeed looks very funky. I think some of the smudges are a noise due to low L and coverage (the one that is on the curve that is L / total_cov). But the smudge that is not on the edge looks like something real, I guess it's hard to say what to make out of it though without more info about the genome / species / isolate.
So Questions:
- Can you also post genomescope? And do you have an expected genome size? Those two are usually very informative in guessing 1n coverage
- Is this a single individual sequenced? The coverage ratio of the nice smudge is 1/6. It looks it's probably missannotated too, it looks more AAAAAB type of locus - which would suggest your coverage is ~21x per haplotype. However, if you would sequence multiple individuals at the same type, it might happen that one haplotype was dominant and the ratio is simply the proportion of the two most frequent genotypes in the pooled sample.
About improving smudgeplot. Currently smudgeplot is not doing great with samples that have wild coverage ratios such as yours, the 1n coverage estimate is quite often wrong. We probably should rethink a bit how we infer the coverage and annotate smudges.
Closing due to inactivity, if naything, please feel free to reopen