ITK
 ITK copied to clipboard
ITK copied to clipboard
Left-handed orientation is discarded when saving a .dcm file
Description
Out of 48 possible orientations for a 3d image, the 24 left-handed orientations are treated incorrectly when saving a multi-frame dicom file or series.
- Examples for right-handed orientations (24 in total):
- RAI
- SAR
- Left-handed (24 in total):
- RAS
- RPI
Steps to Reproduce
- Download nih nifti example: avg152T1_LR_nifti.nii.gz (From: https://nifti.nimh.nih.gov/nifti-1/data) This nifti's orientation is RPI - left-handed.
- Run the following code in python:
import itk
nifti_image = itk.imread('avg152T1_LR_nifti.nii.gz')
nifti_direction = itk.array_from_vnl_matrix(
nifti_image.GetDirection().GetVnlMatrix().as_matrix())
# save as a dicom
itk.imwrite(nifti_image, 'avg152T1_LR_nifti.dcm')
# read the saved dicom
dicom_image = itk.imread('avg152T1_LR_nifti.dcm')
dicom_direction = itk.array_from_vnl_matrix(
dicom_image.GetDirection().GetVnlMatrix().as_matrix())
print(nifti_direction)
# [[ 1. 0. 0.]
# [ 0. -1. 0.]
# [ 0. 0. 1.]]
print(dicom_direction)
# [[ 1. 0. 0.]
# [ 0. -1. 0.]
# [ 0. 0. -1.]]
- Open in itk-snap/slicer the original nifti and then the dicom. The dicom is flipped up-down.
Expected behavior
- The direction matrix should be the same
- The saved dicom image should not be flipped up-down
Actually, 1 leads to 2. I should emphasize that this is not a bug of slicer nor itk-snap. The bug is in our dicom writer.
Actual behavior
- The direction matrices are different. The original direction matrix of the nifti (RPI):
[[ 1. 0. 0.]
[ 0. -1. 0.]
[ 0. 0. 1.]]
The dicom's direction matrix (corresponding to RPS):
[[ 1. 0. 0.]
[ 0. -1. 0.]
[ 0. 0. -1.]]
- In itk-snap, the original nifti is:
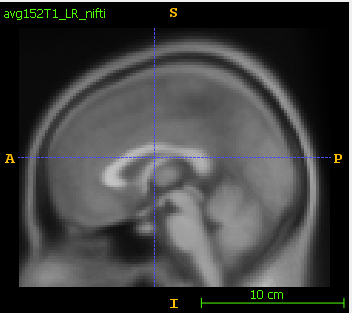 The saved dicom is flipped in S-I axis:
The saved dicom is flipped in S-I axis:
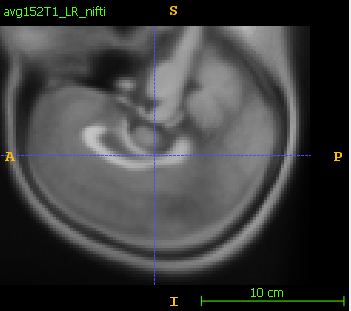
Reproducibility
100% for saving an image with left-handed orientation in multi-frame dicom, both single file (as shown here) and a series of 2d slices. In general, it's relevant for 3d images with left-handed orientations (50% of all possible orientations).
Versions
Itk 5.2.0
Environment
Python 3.9.4, macOS Big Sur (probably irrelevant)
Additional Information
The issue and some possible solutions have already been discussed here: https://discourse.itk.org/t/nifti-rpi-orientation-file-exported-to-dicom-wrong-orientation/1612
Explanation of the bug
The dicom orientation is encoded in the 6 numbers of Image Orientation (Patient) tag.
These 6 numbers correspond to the first and second column of the direction matrix. Let's call these columns u, v (vectors of length 3).
When saving the dicom, the 3rd column of nifti_direction - n, is discarded. When reading the dicom, it is inferred as the cross product of u and v:
- n = u x v
However, in this way it can never be (-n)!
Possible fixes
- Raise an error when saving a dicom with a left-handed orientation. The orientation is easy to check: the determinant of the direction matrix (a rotation matrix) is either +1 or (-1). +1 - right-handed orientation, go ahead (-1) - error.
- As Mihail Isakov suggested, if the image is left-handed - reorient it to the nearest right handed orientation.
- Encode the orientation direction in the sign of Spacing Between Slices tag
+1 for Fix 1.
The following table shows the original orientation of the image in itk vs. the orientation of the saved 3d dicom file (.dcm) corresponding to this image. There are different columns for RGB and non-RGB cases:
| Orientation | Right/Left-handed orientation | Success | Saved orientation | RGB success | RGB saved orientation |
|---|---|---|---|---|---|
| RAI | R | TRUE | RAI | TRUE | RAI |
| IPR | R | TRUE | IPR | TRUE | IPR |
| PLS | R | TRUE | PLS | TRUE | PLS |
| LSP | R | TRUE | LSP | TRUE | LSP |
| ILP | R | TRUE | ILP | TRUE | ILP |
| PIL | R | TRUE | PIL | TRUE | PIL |
| LPI | R | TRUE | LPI | TRUE | LPI |
| SLA | R | TRUE | SLA | TRUE | SLA |
| ASL | R | TRUE | ASL | TRUE | ASL |
| LAS | R | TRUE | LAS | TRUE | LAS |
| IAL | R | TRUE | IAL | TRUE | IAL |
| ALI | R | TRUE | ALI | TRUE | ALI |
| LIA | R | TRUE | LIA | TRUE | LIA |
| SRP | R | TRUE | SRP | TRUE | SRP |
| PSR | R | TRUE | PSR | TRUE | PSR |
| RPS | R | TRUE | RPS | TRUE | RPS |
| SPL | R | TRUE | SPL | TRUE | SPL |
| SAR | R | TRUE | SAR | TRUE | SAR |
| RIP | R | TRUE | RIP | TRUE | RIP |
| ARS | R | TRUE | ARS | TRUE | ARS |
| IRA | R | TRUE | IRA | TRUE | IRA |
| AIR | R | TRUE | AIR | TRUE | AIR |
| RSA | R | TRUE | RSA | TRUE | RSA |
| PRI | R | TRUE | PRI | TRUE | PRI |
| ARI | L | FALSE | ARS | FALSE | ARS |
| LPS | L | FALSE | LPI | FALSE | LPI |
| SAL | L | FALSE | SAR | FALSE | SAR |
| IPL | L | FALSE | IPR | FALSE | IPR |
| PSL | L | FALSE | PSR | FALSE | PSR |
| IAR | L | FALSE | IAL | FALSE | IAL |
| PLI | L | FALSE | PLS | FALSE | PLS |
| LIP | L | FALSE | LIA | FALSE | LIA |
| RAS | L | FALSE | RAI | FALSE | RAI |
| SLP | L | FALSE | SLA | FALSE | SLA |
| IRP | L | FALSE | IRA | FALSE | IRA |
| ALS | L | FALSE | ALI | FALSE | ALI |
| PIR | L | FALSE | PIL | FALSE | PIL |
| LSA | L | FALSE | LSP | FALSE | LSP |
| ASR | L | FALSE | ASL | FALSE | ASL |
| SRA | L | FALSE | SRP | FALSE | SRP |
| ILA | L | FALSE | ILP | FALSE | ILP |
| AIL | L | FALSE | AIR | FALSE | AIR |
| RPI | L | FALSE | RPS | FALSE | RPS |
| RIA | L | FALSE | RIP | FALSE | RIP |
| SPR | L | FALSE | SPL | FALSE | SPL |
| PRS | L | FALSE | PRI | FALSE | PRI |
| RSP | L | FALSE | RSA | FALSE | RSA |
| LAI | L | FALSE | LAS | FALSE | LAS |
Conclusions:
- Saving a non-channeled 3d dicom works only for right-handed orientations (as the original issue suggests)
- ~~Saving an RGB 3d dicom works only for RAI orientation~~
The table can be viewed also here: https://docs.google.com/spreadsheets/d/1ieM8uYYDOBUwoA3_O8FBf7E_72HRb7exE5Sj6KFA0CY/edit?usp=sharing
The code for creating this table: https://gist.github.com/jond01/c77fd1877aa20f6e9ac3cdb8755ec1be
(Edit: sorted the table by R/L.)
Edit: updated the table for ITK v5.2.0. RGB and non-RGB is the same.
@thewtex
So after the issue has been characterized (+-),
I prefer fix 3 with negative Spacing Between Slices because it will not block the user from some orientations.
Regarding the failures in RGB 3d dicom - do you have any idea what is the reason behind that?
@jond01 outstanding investigation! :clap:
Many codes, including ITK, assume that spacing is a physical length that will always be positive, and undefined behavior, which may be wrong, can result when processing, so I think fix 1 is still preferred.
Not sure about the RGB limitation, @malaterre may have ideas.
Spacing Between Slices (0018, 0088) is not taken into account for z-spacing calculation at all in GDCM/ITK in most cases. Long story why, but IMHO it is correct. Most tools rely only on image position (patient) and image orientation (patient) attributes and don't believe anything else.
I've briefly browse through the report, the first thing that pops up is the direct reference to NM module. I must admit I never realized negative value for Spacing Between Slices was acceptable. In my mind this was always like the Enh MR Image Storage:
- http://dicom.nema.org/medical/dicom/current/output/chtml/part03/sect_C.7.6.16.2.html#table_C.7.6.16-2
I'll clarify that GDCM always compute the non-negative Spacing Between Slices in all cases.
Ok, thank you all! So negative Spacing Between Slices is not a possible/adequate way of fixing left-handed orientations. Therefore, I conclude that there should be no left-handed dicoms, leading us to fix 1. I hope to make a PR for this issue in the coming weeks, at least for the non-RGB case.
Another peculiar point which I did not mention - saving RGB dicom series is possible with all right-handed orientations and not only RAI.
Update: The relevant lines in GDCM IO writer: https://github.com/InsightSoftwareConsortium/ITK/blob/27258efa6ba74228958af7b5a620d005c644da51/Modules/IO/GDCM/src/itkGDCMImageIO.cxx#L1000-L1043
Update regarding the 3d RGB case: I dug into the writer's internals - including GDCM - and turns out it's all fine. The direction cosines are set properly, which can be verified with pydicom for example:
import pydicom
ds = pydicom.dcmread('path/to/3d_channelled_ARS.dcm')
orientation = ds.SharedFunctionalGroupsSequence[0].PlaneOrientationSequence[0].ImageOrientationPatient
print(orientation)
# [-0, 1, 0, 1, -0, 0]
Whereas in itk:
import itk
img = itk.imread('path/to/3d_channelled_ARS.dcm')
direction = itk.array_from_vnl_matrix(img.GetDirection().GetVnlMatrix().as_matrix())
print(direction)
# [[1. 0. 0.]
# [0. 1. 0.]
# [0. 0. 1.]]
The expected direction matrix is
[[-0. 1. 0.]
[ 1. -0. 0.]
[ 0. 0. -1.]]
(The first two columns of direction are the orientation in pydicom, the 3rd is their cross product.)
Perhaps, it deserves a different issue and PR as being related to the reader component and not the writer.
I could not reproduce problem with RGB image. Took MHA RGB image in ASL orientation, exported to DICOM (actual ITK master), result is correct - Multi-frame True Color Secondary Capture Image Storage has proper orientation. S. the test and input file (trivial, just read and write). Import DICOM, export MHA seems to work too. May be something was updated, i remember i saw similar issue in the past, but now seems to work well. May be try latest release or master.
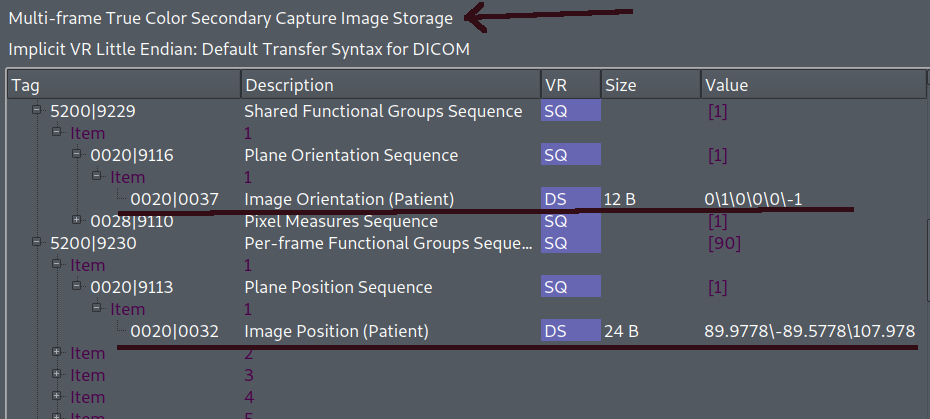
Hi @issakomi, thank you very much for your example and interest! (I used a very similar example to test the RGB images, after changing the number of dimensions to 3.)
I reproduced your example, but there are still two separate issues regarding RGB dicom images:
- Writing:
Not always the image is written with the correct direction cosines (although the SOP class supports it). For example, with your data in python:
The saved dicom is missing the physical data (positions, orientation...). In C++, itk performs the writing well - as you show with Aliza.img = itk.imread('test_rgb/input/t0.mha', pixel_type=itk.RGBPixel[itk.UC]) # the direction matrix is correct: # itk.array_from_vnl_matrix(img.GetDirection().GetVnlMatrix().as_matrix()) itk.imwrite(img, 'test_rgb/input/t0_itkpy.dcm')
The result of your example is here: t0.zip
- Reading: If you open the above zipped dcm file with itk-snap, the orientation is evidently wrong (RAI instead of ASL). Itk's reader seems to ignore the physical data in this case. I suppose Aliza's reader uses bare gdcm and treats the metadata itself?
Yes, itk-snap shows wrong orientation. Probably it was fixed here
https://github.com/malaterre/GDCM/commit/1767b5daae50e29da608ecddd3106ffd90c46bf4#diff-646c7508e8b2ed56649fd3d72843b0a5
This issue has been automatically marked as stale because it has not had recent activity. Thank you for your contributions.
This issue has been automatically marked as stale because it has not had recent activity. Thank you for your contributions.