ontology-development-kit
 ontology-development-kit copied to clipboard
ontology-development-kit copied to clipboard
lost foaf_depicted_by
Hello,
I just updated to ODK 1.3.0. I use dosdp with custom patterns to manage my anatomy ontology. I use foaf_depicted_by. In my most current release I see that I lost these lines in a few different imports and my custom pattern files. Here is a link to my git diffs
http://purl.obolibrary.org/obo/foaf_depicted_by http://purl.obolibrary.org/obo/foaf_depiction
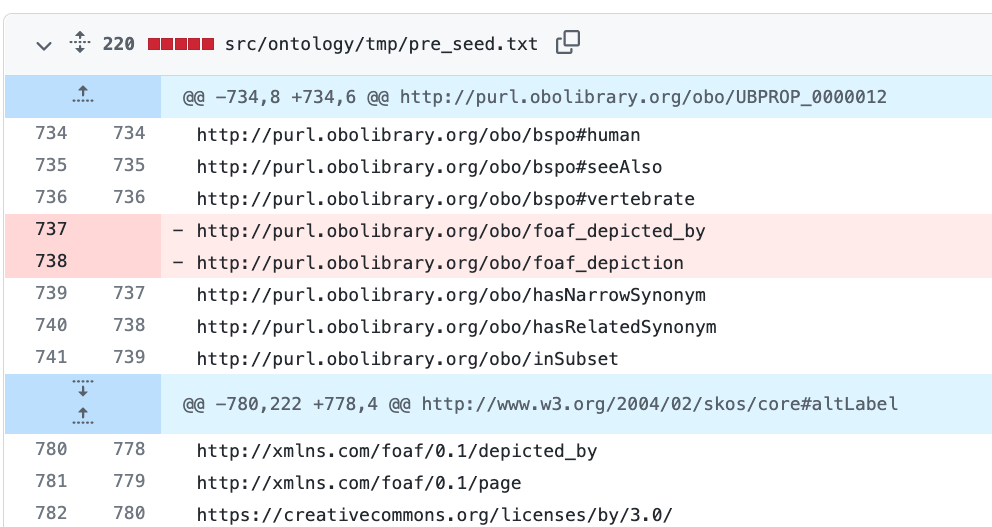
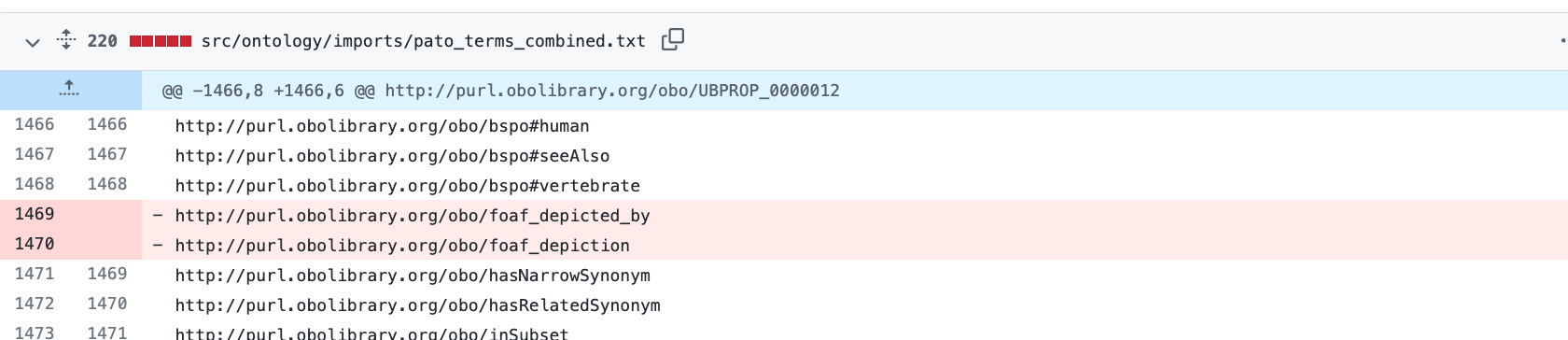
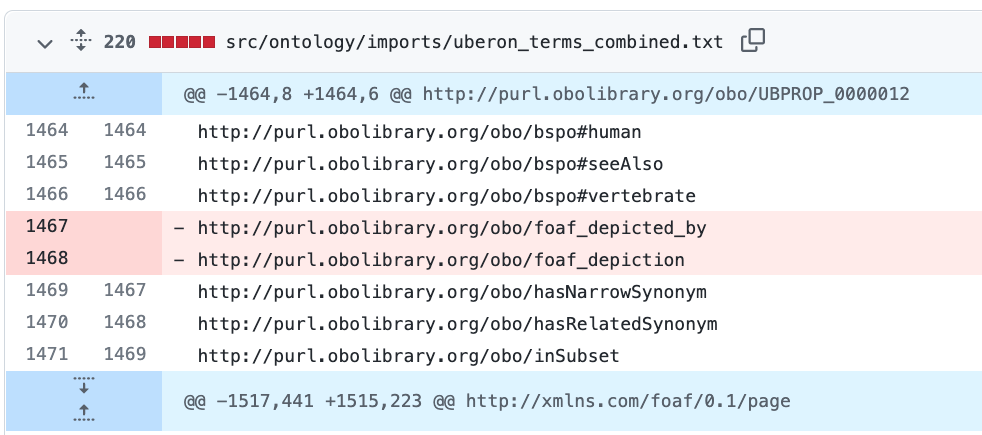
I also lost foaf_depicted_by in my custom pattern as well as nested comment properties.
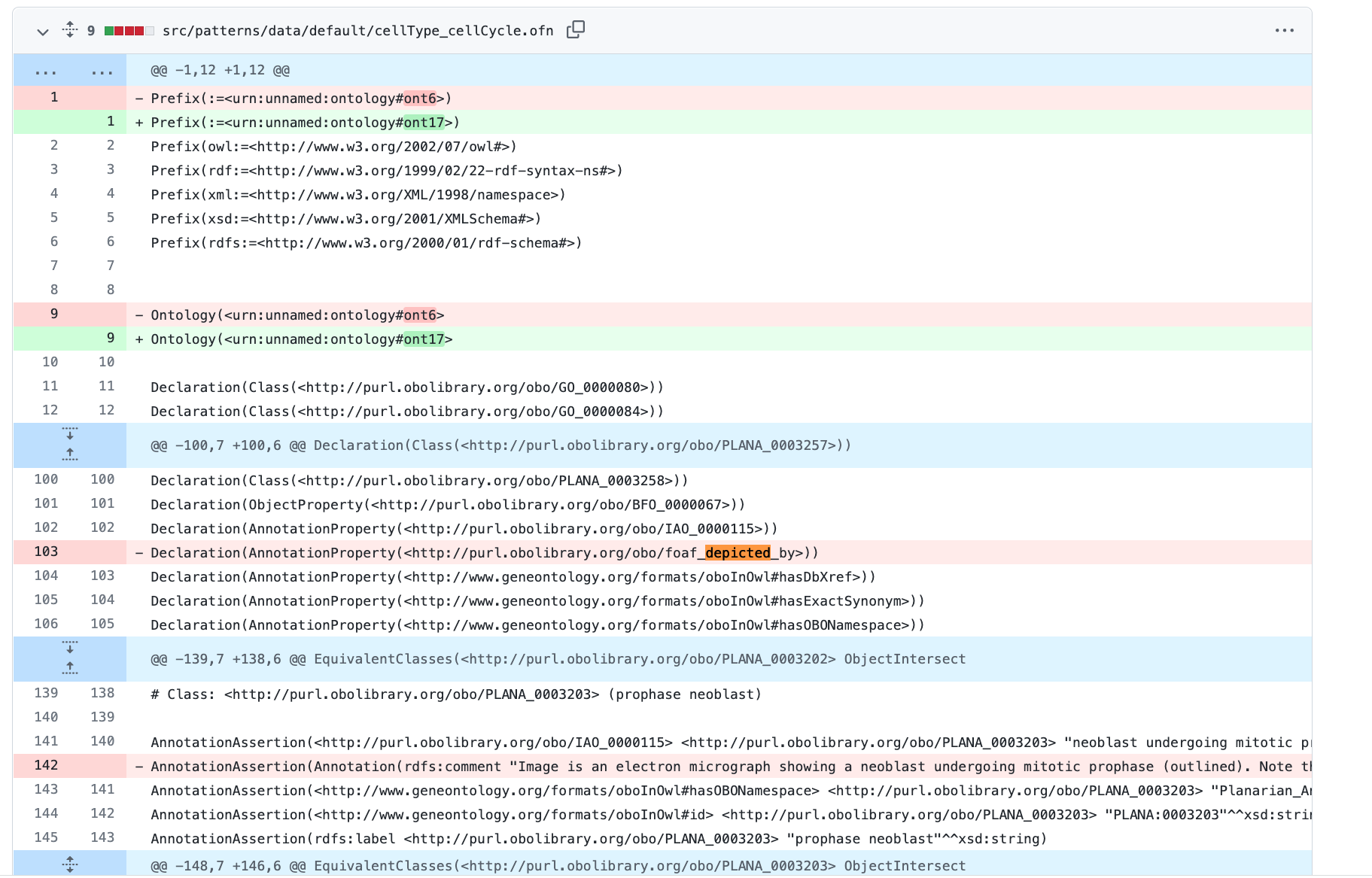
Maybe I just need to change to http://xmlns.com/foaf/0.1/depicted_by
Thanks for any input.
@balhoff does this ring a bell for you? Do you think this is a dosdp tool ticket? Or maybe this is a missing prefix declaration?
I was not sure it would help, but I wanted to try something.
I changed my depicted_by declaration to see if it would help. It did not help at all.
It was:
http://purl.obolibrary.org/obo/foaf_depicted_by
and I changed it to:
http://xmlns.com/foaf/0.1/depicted_by
All my images annotations are still missing.
Sofia
@srobb1 could you point me to how I could run the needed targets in a checkout of your repo?
Hi @balhoff,
Here is my repo: https://github.com/obophenotype/planaria-ontology
I just run sh ./run.sh make prepare_release
if you look in my patterns/dosdp-patterns directory you will see plana_terms.yaml. This includes my depicted_by pattern.
In my data/default/plana.tsv you will see I have data for the depicted_by variable:
| defined_class | defined_class_name | new_entity | new_entity_label | parent_entity | parent_entity_label | exact_syn | defined_class_definition | dbxref | def_dbxref | depictedBydepicted_by | image_comment | comment | has_obo_namespace | created_by |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PLANA:0002060 | nephridiopore | PLANA:0002060 | nephridiopore | PLANA:0002079 | pore | The external opening and terminus of a protonephridium made by a terminal cell. | ISBN:9780070316607 | PMID:21828097 | UBERON:0006972 | ISBN:9780070316607 | https://raw.githubusercontent.com/obophenotype/planaria-ontology/master/metadata/images/nephridiopore.png | Image is an electron micrograph depicting a collecting duct (arrowheads) passing through the basal lamina (BL). Dorsal is up and the scale bar is 5 um. |
This is a problem in dosdp-tools that actually seems to go back a couple of releases. I fixed it and will make a release soon.
Great! Thanks. I guess I have not been using the most up to date dosdp-tools.
@srobb1 when I was debugging this I noticed a few things you might want to change about your pattern YAML:
- The created_by annotation seems to use pipe-separated values but is not set up as a list annotation.
- The values in the
varsdictionary should be the range of the terms in the input. So for something likedepicted_byjust useThing(define this in yourclassesdictionary asowl:Thing). - Should provide a prefix for
foafwhen you run dosdp-tools.
@balhoff
Thanks for the input! And thank you for taking the time to review my yaml, this is extremely helpful. It is super great to get my yamls written properly. I have a couple questions.
created_by
I am using lists but not as in my annotations dictionary. I want to make sure I format this properly,
currently I have my created_by as:
annotationProperties:
depicted_by: foaf:depicted_by
vars:
created_by: "'created_by'"
annotations:
- annotationProperty: created_by
text: "%s"
vars:
- created_by
to change it to a list would it look like this? :
annotationProperties:
depicted_by: foaf:depicted_by
data_list_vars:
created_by: xsd:string
annotations:
- annotationProperty: created_by
value: created_by
depicted_by
These are the changes you suggest. Is the formatting okay for everything depicted_by related? Are my quotes okay in my vars dictionary?
classes:
Thing: owl:Thing
annotationProperties:
depicted_by: foaf:depicted_by
vars:
depicted_by: "'Thing'"
annotations:
- annotationProperty: depicted_by
var: depicted_by
annotations:
- annotationProperty: image_comment
text: "%s"
vars:
- image_comment
foaf prefix
I run ODK prepare_release, how do I provide a prefix for foaf when I run dosdp-tool from within ODK? I have it in my plana-edit.owl Prefix(foaf:=<http://xmlns.com/foaf/0.1/>). Is there somewhere else I need to put it or some other way to handle this?
Thank you again, Sofia
- [x] @srobb1 I think that all sounds right. @matentzn is there a way to pass a
prefixes.yamlto dosdp-tools?
@balhoff I made all the corrections I can (did not do anything about the foaf prefix yet). I changed created_by orcid to use a list. I don't get any errors, but I do now lose all my orcid annotations. Is this due to the same issue I was having that will be corrected with the new release of dosdp-tools?
git diff ../patterns/data/default/plana_terms.ofn
....
# Class: <http://purl.obolibrary.org/obo/PLANA_0000002> (Stage 2)
AnnotationAssertion(Annotation(<http://www.geneontology.org/formats/oboInOwl#hasDbXref> "PMID:28072387"^^xsd:string) <http://purl.obolibrary.org/obo/IAO_0000115> <http://purl.obolibrary.org/obo/PLANA_0000002> "A stage of Smed embryonic development defined by a unique gene expression signature and morphology, 2 - 3 days post-egg capsule deposition at 20˚C. Sphere formation. A fraction of the blastomeres differentiate into temporary embryonic tissues that provide form and function to the embryo. Undifferentiated blastomeres remain in the embryonic wall."^^xsd:string)
-AnnotationAssertion(<http://www.geneontology.org/formats/oboInOwl#created_by> <http://purl.obolibrary.org/obo/PLANA_0000002> "https://orcid.org/0000-0003-2569-1939 | https://orcid.org/0000-0002-7535-4407"^^xsd:string)
Thanks, Sofia
@balhoff I made all the corrections I can (did not do anything about the foaf prefix yet). I changed created_by orcid to use a list. I don't get any errors, but I do now lose all my orcid annotations. Is this due to the same issue I was having that will be corrected with the new release of dosdp-tools?
My guess is yes.
Sorry for the long wait on this: it was possible all along using the dosdp_tools_options option in the config file! You can pass any additional dosdp parameters through there! If this for some reason does not work for you, please reopen and let me know!