Proposal: use UCUM for units across all of MIxS
I'm carrying over some of my comments from https://github.com/GenomicsStandardsConsortium/mixs/issues/93#issuecomment-811541452
That ticket is about whether units should be baked into each column. My proposal here applies to MIxS regardless of the outcome of decisions on issue 93
Backgound: MixS current recommends units are spelled out, e.g. "meter"
I recommend revising this and using UCUM unit codes as standard. UCUM: https://ucum.org/ucum.html
For 'meter' the UCUM code is m
UCUM largely follows SI and has been adopted by many standards orgs, eg HL7/FHIR
Note there is also a collaboration between OBO, UO, OE, and BCODMO to come up with a semantic web standard for units based on UCUM (cc @kaiiam). Here is a prototype resolver, https://units.ontodev.com/m
In addition to following standards, I recommend this as full names 'meter' and the like will lead to orthographic errors e.g. meters, metres, metre, meter. It is not very international as other languages will have different labels.
A possible objection to my proposal is that it is a breaking change with current mixs. However, if we look in INSDC at how MIxS has been used, it seems that most people have ignored MIxS recommendations here anyway. E.g I see ~3k samples have a depth field of the form number m; ~1k of the form number cm. In contrast there are ~300 samples using some variant of (meter|meter)[s]
The new system created in collaboration with James Overton, Simon Cox and many others is intended to bridge the gap between existing systems UO, QUDT etc by leveraging UCUM, and as @cmungall said creating a semantic web ontology wrapper for it. The system takes UCUM codes as input and creates ontology terms based on the codes.
for example m/s currently outputs the following:
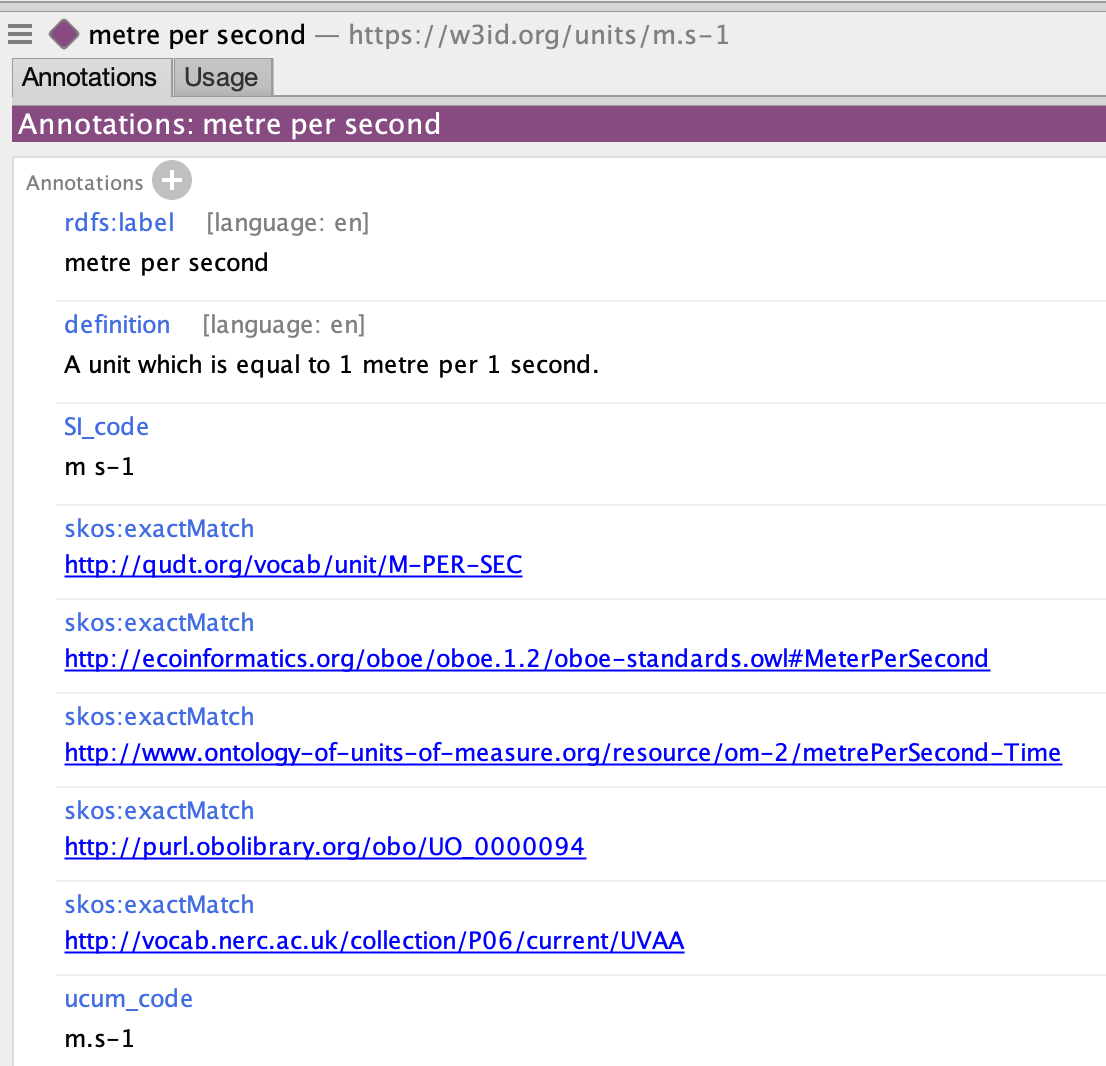
Once the server is setup this will be a dynamic process, but for now you can checkout the current version of the generated ontology here.
I think @cmungall's suggestion of having MIxS use the UCUM codes is good because the codes can easily be put through this system to get IRIs.
Alternatively, the new system includes skos matches to IRIs from other Unit systems, so if there were a set of MIxS IRIs which you generated from the current labels used in MIxS (e.g. https://mixs_prefix..._meter we could map those to UCUM and then add them to the new scripts mapping layer (like we do for QUDT, UO, etc).
A third option is to use NLP to label match to the new systems labels and (soon to be added) synonyms. We plan to automate the generation of synonyms to cover cases like meters, metres, metre, meter.
Hope this helps.
I think using NLP/text matching is a great solution for people who are looking to normalize messy data (in fact we are doing this in a project where we are trying to semi-automatically tidy all data in INSDC biosample)
However, I think the recommendations that MIxS provides should be very clear and simple and unambiguous
Agreed hence my support for MIxS using the UCUM codes, they can later be resolved to the new ontology IRIs.
On Tue, May 25, 2021 at 8:48 PM Chris Mungall @.***> wrote:
External Email
I think using NLP/text matching is a great solution for people who are looking to normalize messy data (in fact we are doing this in a project where we are trying to semi-automatically tidy all data in INSDC biosample)
However, I think the recommendations that MIxS provides should be very clear and simple and unambiguous
— You are receiving this because you were mentioned. Reply to this email directly, view it on GitHub https://github.com/GenomicsStandardsConsortium/mixs/issues/154#issuecomment-848158851, or unsubscribe https://github.com/notifications/unsubscribe-auth/AC5QDSDZQJWMXXBLPZ2AH43TPPWH5ANCNFSM45JY27RQ .