jbrowse-components
 jbrowse-components copied to clipboard
jbrowse-components copied to clipboard
Optimize SNPCoverage rendering on large regions
I mentioned recently that arc view would sort of put pressure on our code to optimize other parts of our system, since arc track is often useful to view in a zoomed out view. One of those parts of the system is snpcoverage, which is useful to display alongside arc track to see e.g. a drop in coverage
here is a a 600kb region with ~100x illumina reads from a BAM file. not small data, but it is useful to render everything as we can see it
the above screenshot takes about a minute to render, with ~1/2 of the time occupied by generateCoverageBins (figure from speedoscope with chrome profiler)
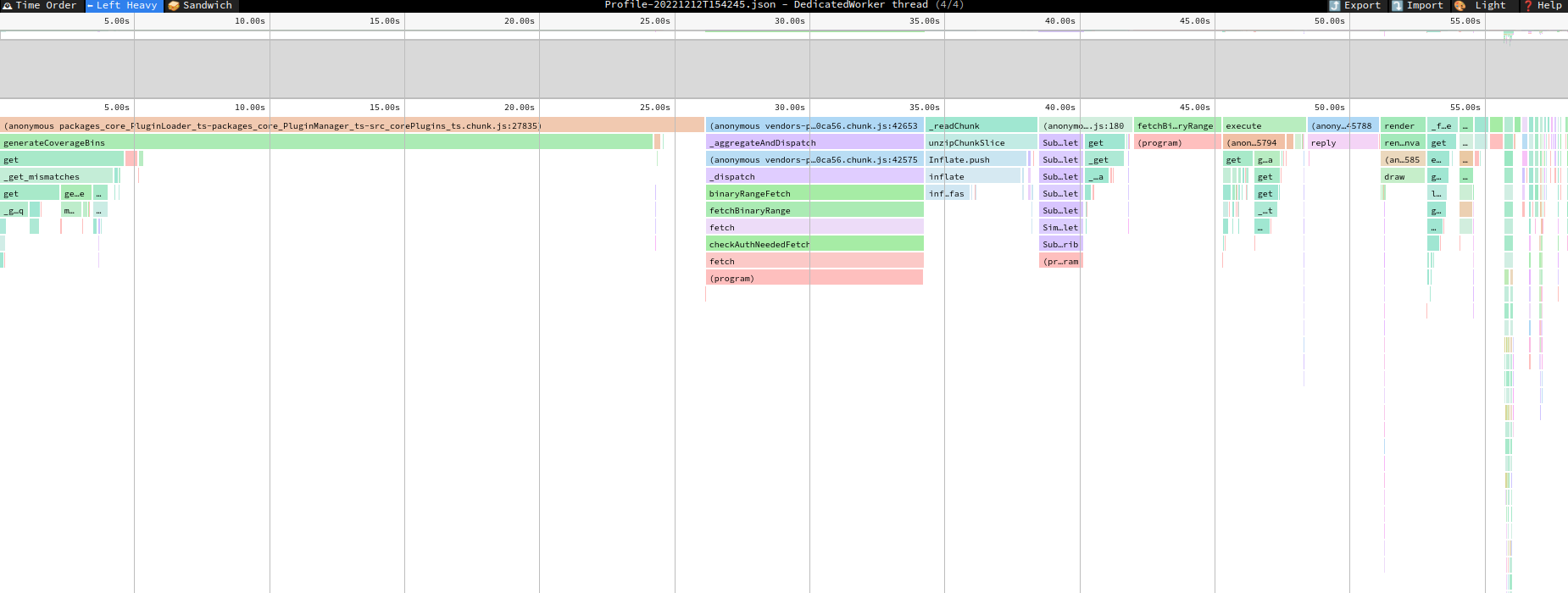
indeed, the code would need to allocate an array of 600,000 objects for snpcoverage because we don't perform binning. we could try to figure out some binning though and it may improve performance
possibly related https://github.com/GMOD/jbrowse-components/issues/2814 at least, it is affected because if you scroll a little, it can cause a big minute long re-render
Tried looking at a ~1Mbp region of RNA-seq data w/ @carolinebridge-oicr and it was very slow. The code likely needs to 'bin' the data instead of per-base statistics for the coverage as 1Mbp is a million little javascript objects with more nested javascript objects, gets slow
The reason for zooming out to this large area was that the RNA-seq data was very sparse, and trying to find 'interesting data' in region. We could also consider 'semantically changing to e.g. a bigwig or indexcov' at that scale, but i think the js could still be optimized
see https://github.com/brentp/mosdepth#how-it-works as a possible method to optimize our snpcoverage.
for example, the pseudocode way the way our code currently works is:
for(const read of reads):
for(const position in read.start...read.end):
increment bin[position]
therefore, if the average read length is 100bp, and the number of reads is N, then the runtime is 100*N
the mosdepth algorithm only increments and decrements at the start and end of the read so it's like 2*N, which is probably a substantial speed up
can look at http://jbrowse.org/code/jb2/main/?config=test_data%2Fconfig_demo.json&session=share-1qNYykeMaS&password=cg9jv to see long read rna-sequencing (e.g. iso-seq but nanopore) and it is fairly performance intensive, would be good to optimize and a lot of the time is spent on generateCoverageBins from snpcoverage
(link above not yet valid, but will be once site is deployed)