Bring FAN into CTSM
We want to bring @juliusvira FAN (Flow of Agricultural Nitrogen) model into CTSM. @wwieder. Before creating a PR for it, this issue can report on this task.
Here's the initial implementation:
https://github.com/ESCOMP/ctsm/compare/clm5.0.dev003...juliusvira:fan-2019
As an initial request for a change: The "ndep2" and "ndep3" streams files need better names.
ndep2 was renamed, and ndep3 was removed.
@negin513 and @billsacks note the following.
There are some CPP #ifdef's and that's something we've tried to move away from. There's also a bunch of constants that are set in FAN, and the global constants from CESM and CLM should be used. One way to handle that, would be to have them available for offline use, but overridden by the host land-model for consistency. So we need to talk about that as well. It looks like the CPP tokens are there so that you can run this from Python, that might be an exception we would allow, or at least figure out a way to do it that's acceptable to everyone. I do certainly condone the usage of running this standalone from Python! That's an ability I wouldn't want you to loose. And it's actually the kind of thing that I think would be good to endorse for other parts of the code. I'm betting that it would be very very very useful for scientists to be able to run parts of the code from Python to check specific things out.
@negin513 and I talked with @juliusvira yesterday. We came up with some thing's we'd like to see:
- Move shared parameters to module data
- Remove the CPP token that's just there for r8 kind, as we don't envision using a FORTRAN standalone driver
- Add more comments on what's going on. Look at other parts of CLM for formatting. (also see: https://wiki.ucar.edu/display/ccsm/CLM+Coding+Conventions)
- Add a FANtoCTSM module to interface to CTSM specifically, so that FAN can be separable from CESM.
- Add a Nitrogen balance check for the FAN pools, to ensure FAN is conserving Nitrogen. This can be called when FAN is run.
- I think there are still parameters that should use the CESM/CLM constant value rather than FAN's particular set (for example Tref).
Some things we need to look into is the use of F2Py versus some other method to be able to import FAN into Python. This is a useful functionality that we want to keep (and to have the capability to be used for other parts of CTSM). Depending on the outcome of that, we might have different suggestions on handling the CPP tokens.
I have tried to address these ideas in the new branch https://github.com/juliusvira/ctsm/tree/fan-2019-ctsm:
- Added the Fan2CTSMMod module as an interface between FAN and the rest. This also reads the parameters from namelist.
- Added the FAN pools and fluxes to the N balance check. The totals are first aggregated by FAN to keep the changes small elsewhere.
- Changed FAN to use the SHR-constants if possible
- Removed the r8 CPP token
- Added some comments
There is another issue with the python interfacing, namely, the current f2py does not support assumed shape arrays (arrays declared with (:)). FanMod now uses old-style subroutine calls with explicit arguments for array dimensions. This is not ideal, but previously many array sizes were hard coded, which probably was even less robust. The next release of f2py is supposed to support assmed shape, but it's not yet out.
This version also includes the option to connect FAN with the CLM nitrogen cycle.
@juliusvira and @ekluzek where are we with this? how close are we to being able to bring FAN into CTSM/CLM?
@juliusvira and I met to talk about the code. Going over it I think most of the changes to the code have been done. Still some constants could be worked on. Some constants are declared in multiple subroutines and could be module data for FAN. It's currently up to date with ctsm1.0.dev026, so we need to update it to the newest version. We also need to make sure it's default off and namelist variables to turn on, and then we need to add some tests for it on as well. We also need input files for it. Then we'll need to do a PR to bring it to CTSM master. So we also need to decide on who is going to do what on each of these things here.
@juliusvira it's tricky to evaluate some of the simulations you've done because it doesn't look like they're done from an identical code base. As a result Fields like TLAI are not identical, where they may be expected to be? (e.g. R0_CTSM_FANON - R0_CTSM_NOFAN see /glade/scratch/wwieder/fanon-nofan_2011.nc) .
To address this can you clone the fanon simulations and setting name list options (as below, or based on pull request) https://github.com/ESCOMP/ctsm/blob/0865091a5176c55bb51d7efc2715607bd3183608/bld/namelist_files/namelist_defaults_ctsm.xml#L454
This could be done by creating an identical series of simulations (2009-2012) in /glade/u/home/juliusv/ctsm/cases/ that are modified by changing settings in user_nl_clm: 1a. use_fan=.false. ! This should give a ‘nofan simulation’ 2a. use_fan=.true. & fan_to_bgc_crop = .false. ! This should give a ‘fan simulation’ 3a. use_fan=.true. & fan_to_bgc_crop = .true. ! This should give a ‘fanbgc_crop simulation’ 4a. use_fan=.true. & fan_to_bgc_crop = .true. & fan_to_bgc_veg = .true. ! This should give a ‘fanbgc_full simulation’
alternatively maybe this should be done in user_nl_clm with: 1b. fan_mode = .false. 2b. fan_mode = .on. ! should be like 2a, abive 3b. fan_mode = .soil. ! I'm not sure what this does, it it a 'fanbgc_crop' run? 4b. fan_mode = .full. ! what does this do? Will it try and pass fluxes to the atmosphere?
@ekluzek, what's the best way for us to use name list changes to test what's going on?
@juliusvira & @ekluzek I would assume that option 1 should not generate answer changes from simulations done with the base (master) branch? Is this unrealistic?
In addition it may be helpful to turn on some column level output so we can see what’s going on For all of the simulations you listed, ! h1 stream (monthly average, landunit-level) hist_mfilt(2) = 1 hist_dov2xy(2) = .false. hist_nhtfrq(2) = 0 hist_type1d_pertape(3) = 'LAND' hist_fincl2 += 'SMINN', 'FERT_TO_SMINN', 'MANU_TO_SMINN', 'GPP', 'TLAI', etc… any other column-level output that may be helpful…
I have now made a bunch of test runs, history files in /glade/scratch/juliusv/archive/:
- fan2019-nofan from codebase without FAN modifications
- fan2019-fanoff (use_fan = .false.)
- fan2019-fanon-nosprd (use_fan = .true. and fract_spread_grass = 0.0 for consistency with the bgc runs)
- fan2019-fanbgc-crop (fan_to_bgc true for crop)
- fan2019-fanbgc-full (fan_to_bgc true for both crop and native vegetation)
There are some (small but above numerical precision) local differences in TLAI and GPP between fanon and nofan. The global totals are identical up to 5 digits.
Some examples from the results:
Going from fanon to fanbgc-crop changes TLAI for crop columns:
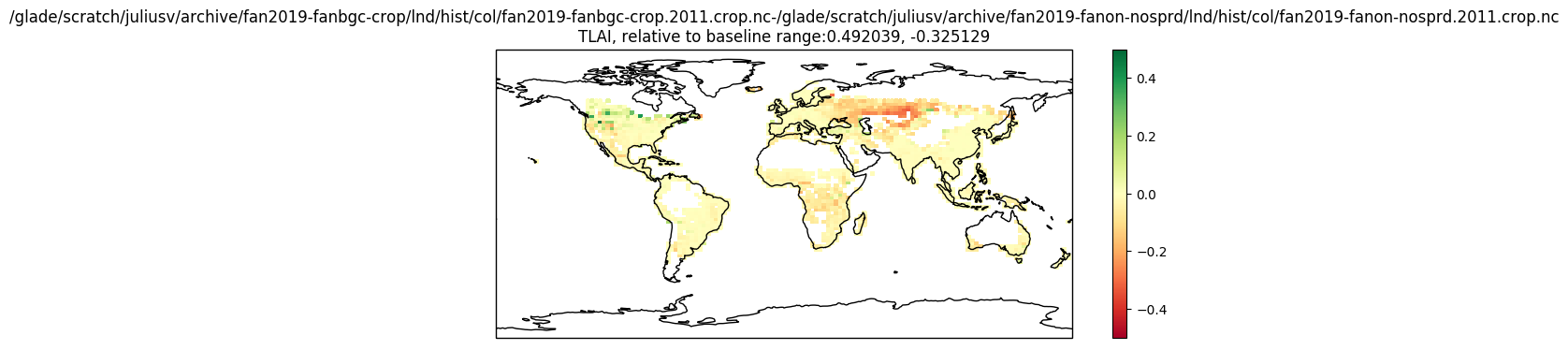
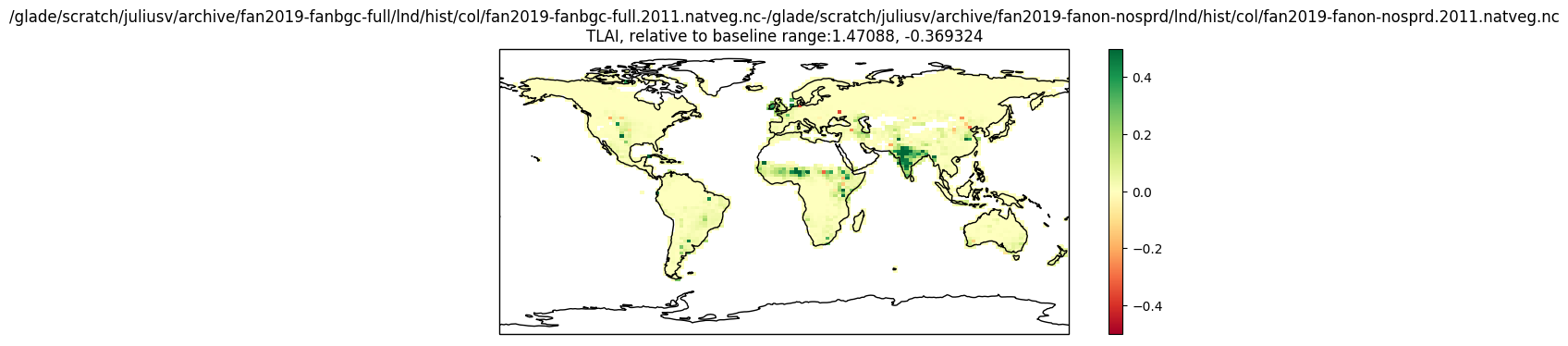
Coupling also nat veg columns (grazing) changes natural veg columns' TLAI substantially in some regions:
I have also checked the some mass budgets from the history files:
- integral FAN_TOTNIN-FAN_TOTNOUT is equal to change in FAN_TOTN
- integral of FAN_TOTNIN - NH3_TOTAL - FERT_NH4_RUNOFF - MANURE_NH4_RUNOFF is equal to FERT_TO_SMINN + change in FAN_TOTN for the columns that are coupled to bgc.
These checks match to numerical precision for all months except January. The discrepancy turns out happen on January 1st each year. I am thinking that this is due to dynamic land use in a transient run, can someone confirm this?
Most of the code review comments should be handled in the latest commits. I also removed the legacy state and flux variables.
Thanks for doing all these runs, Julius. I'll have a closer look at some of the details in advance of our call.
On Tue, Sep 10, 2019 at 6:11 PM juliusvira [email protected] wrote:
I have now made a bunch of test runs, history files in /glade/scratch/juliusv/archive/:
- fan2019-nofan from codebase without FAN modifications
- fan2019-fanoff (use_fan = .false.)
- fan2019-fanon-nosprd (use_fan = .true. and fract_spread_grass = 0.0 for consistency with the bgc runs)
- fan2019-fanbgc-crop (fan_to_bgc true for crop)
- fan2019-fanbgc-full (fan_to_bgc true for both crop and native vegetation)
There are some (small but above numerical precision) local differences in TLAI and GPP between fanon and nofan. The global totals are identical up to 5 digits.
Some examples from the results:
Going from fanon to fanbgc-crop changes TLAI for crop columns: [image: 1991-2011] https://user-images.githubusercontent.com/31929342/64658553-e1385080-d405-11e9-9620-ddbdca7d9a43.png
Coupling also nat veg columns (grazing) changes natural veg columns' TLAI substantially in some regions: [image: 1991-2011] https://user-images.githubusercontent.com/31929342/64658589-062cc380-d406-11e9-8353-b3281937415f.png
I have also checked the some mass budgets from the history files:
- integral FAN_TOTNIN-FAN_TOTNOUT is equal to change in FAN_TOTN
- integral of FAN_TOTNIN - NH3_TOTAL - FERT_NH4_RUNOFF - MANURE_NH4_RUNOFF is equal to FERT_TO_SMINN + change in FAN_TOTN for the columns that are coupled to bgc. These checks match to numerical precision for all months except January. The discrepancy turns out happen on January 1st each year. I am thinking that this is due to dynamic land use in a transient run, can someone confirm this?
Most of the code review comments should be handled in the latest commits. I also removed the legacy state and flux variables.
— You are receiving this because you were mentioned. Reply to this email directly, view it on GitHub https://github.com/ESCOMP/ctsm/issues/636?email_source=notifications&email_token=AB5IWJCNYY4LK22636I27WTQJAZTLA5CNFSM4GXYTOBKYY3PNVWWK3TUL52HS4DFVREXG43VMVBW63LNMVXHJKTDN5WW2ZLOORPWSZGOD6M3ITY#issuecomment-530166863, or mute the thread https://github.com/notifications/unsubscribe-auth/AB5IWJEALQLLQ5ULYBNMC63QJAZTLANCNFSM4GXYTOBA .
-- Will Wieder Project Scientist CGD, NCAR 303-497-1352
See https://github.com/ESCOMP/CTSM/projects/24 for details on what needs to be done.
It's been hard to find the time, but I have by now fixed and tested most of the smaller requests. I still need to change the "take-the-first-found" code to be independent of the column order.
It seems to me that fixing the loop order would be best done by splitting the handle_storage subroutine into two: the first part would distribute the input N between columns, and the second part would evaluate the emission in a simple column loop. This would also avoid the awkward transfer of N between columns, and isolate (if not entirely remove) the checks for column weights. Unfortunately I don't think I will be able to do this in the near future.
Thanks for the update, Julius. it would be good to try and wrap this up, as another group may be working on a somewhat redundant project & could benefit from getting to focus on passing fluxes to the atmosphere.
On Fri, Jan 31, 2020 at 7:58 AM juliusvira [email protected] wrote:
It's been hard to find the time, but I have by now fixed and tested most of the smaller requests. I still need to change the "take-the-first-found" code to be independent of the column order.
It seems to me that fixing the loop order would be best done by splitting the handle_storage subroutine into two: the first part would distribute the input N between columns, and the second part would evaluate the emission in a simple column loop. This would also avoid the awkward transfer of N between columns, and isolate (if not entirely remove) the checks for column weights. Unfortunately I don't think I will be able to do this in the near future.
— You are receiving this because you were mentioned. Reply to this email directly, view it on GitHub https://github.com/ESCOMP/CTSM/issues/636?email_source=notifications&email_token=AB5IWJAELDTALVH6I5IJDITRAQ4DDA5CNFSM4GXYTOBKYY3PNVWWK3TUL52HS4DFVREXG43VMVBW63LNMVXHJKTDN5WW2ZLOORPWSZGOEKO453Y#issuecomment-580767471, or unsubscribe https://github.com/notifications/unsubscribe-auth/AB5IWJFZHGU3GW2PHS2RBYDRAQ4DDANCNFSM4GXYTOBA .
-- Will Wieder Project Scientist CGD, NCAR 303-497-1352
I have now pushed a set of changes trying to address Bill's comments as much as I can at this point. I have now removed the dependency on pft/column ordering. Based on one-year the output seems consistent (within numerical precision) with the previous version. If there will be a need for "final" test runs I can still do them. We could have a telecon on this if it seems useful.