Introduce time-evolving LEAFCN_TARGET as function of leafcn parameter
Description of changes
From clm_driver I call new subr. time_evolv_params located in CanopyStateType.F90 where I calc. the new variable leafcn_patch (history field LEAFCN_TARGET).
Specific notes
Contributors other than yourself, if any: @wwieder @ekluzek
CTSM Issues Fixed (include github issue #): #1646
Are answers expected to change (and if so in what way)? Yes, once I replace all pftcon%leafcn with canopystate_inst%leafcn_patch(p)
Any User Interface Changes (namelist or namelist defaults changes)? New history field LEAFCN_TARGET
Testing performed, if any: In /glade/scratch/slevis, both of these tests PASS: SMS_Ly3_Mmpi-serial.1x1_numaIA.IHistClm50BgcCropQianRs.cheyenne_intel.clm-default.20220218_111830_bodlmh SMS_Ly3_Mmpi-serial.1x1_numaIA.I2000Clm50BgcCropQianRs.cheyenne_intel.clm-default.20220218_105852_zphwfb I will plot timeseries of LEAFCN_TARGET to compare the IHist case to the I2000 case. For now I have only looked at a single hist. file (SMS...I2000...clm2.h0.2002-02-28-00000.nc) and found: leafcn_target = 26.0 leafc / leafn = 55.4 leafcn = 34.7 (Leaf CN ratio used for flexible CN)
Hi Sam,
Thanks for getting started on this. I don't really understand what you're reporting with this test, but I don't understand why leafcn_target < leafcn given the code mods you made? Even more surprising is the high values for leafc / leafn. Maybe there are some details here to discuss?
leafcn_target = 26.0
leafc / leafn = 55.4
leafcn = 34.7
The time series may clarify... @wwieder the high C:N may have been specific to the model day (Feb 28) that I randomly looked at.
1x1_numa_I2000
1x1_numa_IHist
As I replace leafcn(ivt(p)) (the parameter) with leafcn_patch(p) throughout the code, I now see that it makes even more sense to declare leafcn_patch in CNVegNitrogenStateType.F90 rather than in CanopyStateType.F90. I say this because the model uses leafcn_patch in CNVegNitrogenStateType.F90 right after reading the restart file.
If I make this change, FATES will not have access to the time-evolving leafcn, but that probably doesn't matter while FATES is C-only. @wwieder and/or @ekluzek pls confirm, thanks.
Moving it to CNVegNitrogenStateType makes sense to me, as it's really a BGC variable and not a canopy variable that's always used (including in SP mode). By appearing in CanopyState you get the impression that it's also used in SP mode and it's not. You are right that means it's not available to FATES, but I think for the purposes you are using LEAFCN_TARGET it's OK that it won't be available with FATES. I don't think FATES has a similar variable that it could operate on, so this whole thing would need to be rethought for working inside of FATES. And you are right the main FATES code is Carbon only, with nutrients coming in as an extension. So Nitrogen is being worked on, but it's new development.
Sanity check after moving leafcn_patch from CanopyStateType to CNVegNitrogenStateType (I will push corresponding commit soon):
Got bfb identical history from SMS_Ly3_Mmpi-serial.1x1_numaIA.IHistClm50BgcCropQianRs.cheyenne_intel.clm-default as before.
@wwieder mods were more widespread than I had originally expected, but I think I got them all.
This far, testing tells me that the new code is working.
How would you like to proceed:
- Review the code?
- Suggest some runs? Longer and/or global?
wow, there were a lot of changes @slevisconsulting Thanks for doing all of this.
Can we try a few fully transient single point historical runs (maybe for Harvard Forest, 42.5° N, 72.2° W, where we have a good foliar C:N record).
- Let's do this with
co2_base = 310, which means foliar stoichiometry will start changing around 1940. - It's likely easiest to have just a single pft for this run (temp decid tree, 7) and no LULCC.
- A 400 y AD spinup and 200y postAD for 1850 would be enough, unless you can interpolate from initial conditions.
- Then let's do two historical simulations (starting in 1850). One each with:
cn_slope = 0, which should give identical results to the current parameterization andcn_slope = 30, so we get a pretty big change.
cn_slope = 0 during spinup 400 yrs AD_SPINUP transient CO2 and climate but nothing else use subset_data to get fsurdat question about whether will need mesh domain file for this 1x1 site ./create_newcase with same pts_lat and pts_lon that I select in subset_data and don't mess w datm streams (OR ./run_neon for harv but then I have to change datm streams but don't have to create fsurdat)
Ran ./subset_data.py point --site harvard_forest --create_domain True --create_landuse True
A couple of comments:
- I didn't specify lat, lon, dompft and some other things because subset_data defaults to the values that we wanted.
- I set create_domain True in case the model asked for the domain file (it didn't).
- I set create_landuse True because Hist compsets require a landuse file. Then in user_nl_clm I set:
do_transient_pfts = .false.
do_transient_crops = .false.
I have now started the ad_spinup simulation.
I wanted to thank @negin513 and @adrifoster for making subset_data a pleasure to use.
Thanks for this feedback @billsacks. Before spending any time on this @slevisconsulting I think we need to decide if this is a feature we want to include in the model, or if it's a one-off for the project that Emma's doing. We'll be discussing the science on this PR at the CLM meeting next week.
Note that https://github.com/ESCOMP/CTSM/pull/1705 (which I plan to merge soon) will result in more conflicts with this PR.
To extend the simulations (cn_slope_0 & cn_slope_30) to 2100:
- I am first repeating the 1850-2014 period because I originally cycled the 1901-1920 atmosphere data through the entire simulations. Now I will stop the cycling in 1920.
- From 2015 to 2100 I will use SSP3-7.0 anomalies as explained here.
Sam, I'm not sure we need single point runs here, but I actually did the global transient runs. I wonder if I should also extend these for the SPP? /glade/work/wwieder/ctsm/flexLeafCN/cime/scripts
To make this even more complicated, I used mct for the historical simulation (and source mods to set cn_slope = 20).
@emhauser and I have also been discussing putting an upper limit on the changes in leafcn_patch that we're calculation, but haven't finalized what we want to do here.
Sam, I'm not sure we need single point runs here
I canceled those.
One comment on the current approach we're taking.
Under the SSP3-70 scenario co2 gets to ~1000 ppm by 2100. with the current "additive" parameterization all pfts will have their targetCN increase by 23.
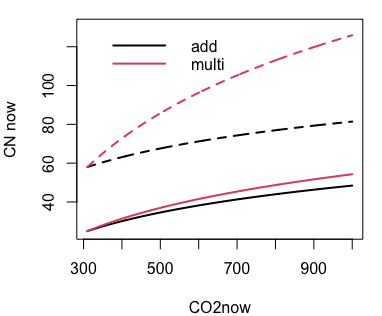
This would be a nearly doubling of foliar C:N for a PFT that's parameterized with a target C:N of 25 (deciduous forests?, solid), but only a 40% increase for a PFT with a target C:N of 58 (evergreen boreal forests, dashed), black lines below.
This makes me wonder if we should use a different function that scales the % change in C:N? For example: cnMulti = 1+log(CO2now/CO2base) This gives just over a doubling of leafCN, shown in the red lines below. We can figure out ways to modify this formula, but it gives pretty different results for PFTs that start with high C:N values.
From what you've seen in the data, is there any way to tell which approach may be more correct Emma?
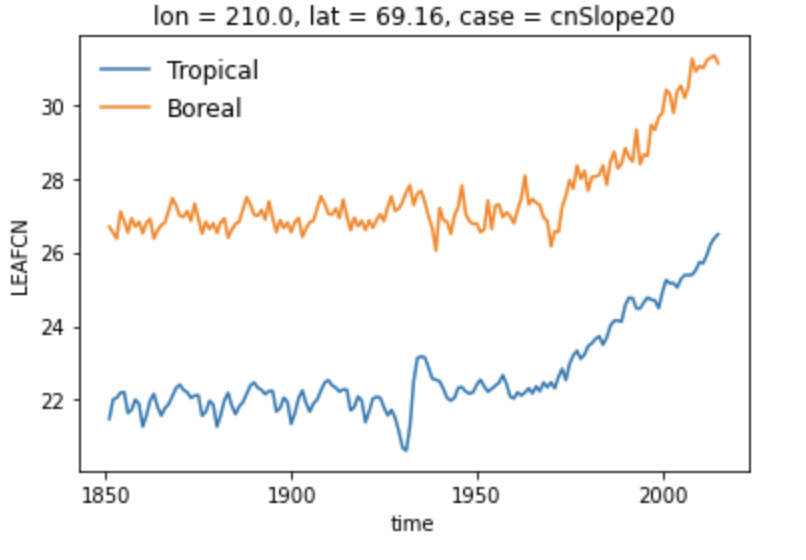
The model data from the global runs through 2015 are like the "add" lines in black above:
My sense from reading is that % change in foliar C:N (and the multiplicative model) is more likely than additive change although the % change varies somewhat by ecosystem. This plot from Du et al (2019) reports % change in C:N between ambient and elevated CO2 scenarios for several different types of ecosystem :
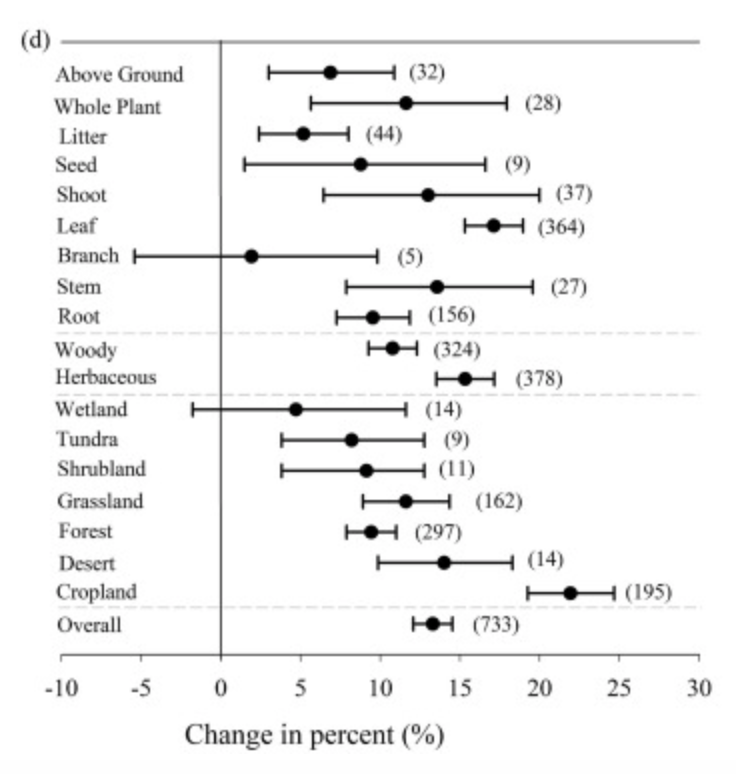
Based on this it doesn't seem like the % change in CN is all that different among systems although wetlands are a little lower and grasslands/croplands a little higher (although error bars overlap in all cases). Based on this it seems like we might want to have a comparable % change across systems rather than an absolute value. I'm still looking around for other helpful datasets but maybe this could be a starting place?