picongpu
 picongpu copied to clipboard
picongpu copied to clipboard
Update binary collisions, dynamic Coulomb log
Update to the binary collisions
This PR introduces a dynamic calculation of the Coulomb logarithm for binary collisions. It also includes some fixes for the existing implementation. These changes continue to follow the implementation from smilei. @ComputationalRadiationPhysics/picongpu-maintainers @psychocoderHPC @HighIander
Dynamic Coulomb logarithm
The newly introduced automatic Coulomb log calculation is based on the 2012 paper from Perez link. The calculation uses a Debye length that is calculated on every time step from all charged particles (the list of included species is set in collision.param) for all simulation cells. These values are made available to the colliders via a FieldTmp. The collision functor has now two implementations: one for a constant and one for a dynamic Coulomb logarithm. More details are in the updated documentation.
Other updates
- A factor of two difference in the s-parameter between the inter- and intra-collisions was removed. Originally, this difference comes from the 2020 Higginson paper link, but it seems to be a typo. This factor two is not present in smilei.
- The sampling of the cosine of the cumulative scattering angle was updated to follow the new fit recently introduced in
smilei. - A debug output was introduced. It can write average values (averaged over the simulation) for the coulomb log, the s-paramer (proportional to the collision frequency) per collider and species pair. It is also possible to write an average value of the precalculated Debye length. It would be probably useful to be able to write these value per cell, but, for now, the averaging over the simulation is good enough for the homogenous test cases.
Tests
The existing collision tests setups were extended to include the new automatic coulomb logarithm. They were also run for a longer time and include now a direct comparison with smilei values and some statistical analysis. Also, the tests use now only field output, so they run faster. A python tool for generating the comparison plots was also included for both tests.
The test results show in general quite good agreement with smilei but there still seems to be some difference in the dynamic debye length and the coulomb log, for example in this beam relaxation set-up:
beam relaxation example set-up
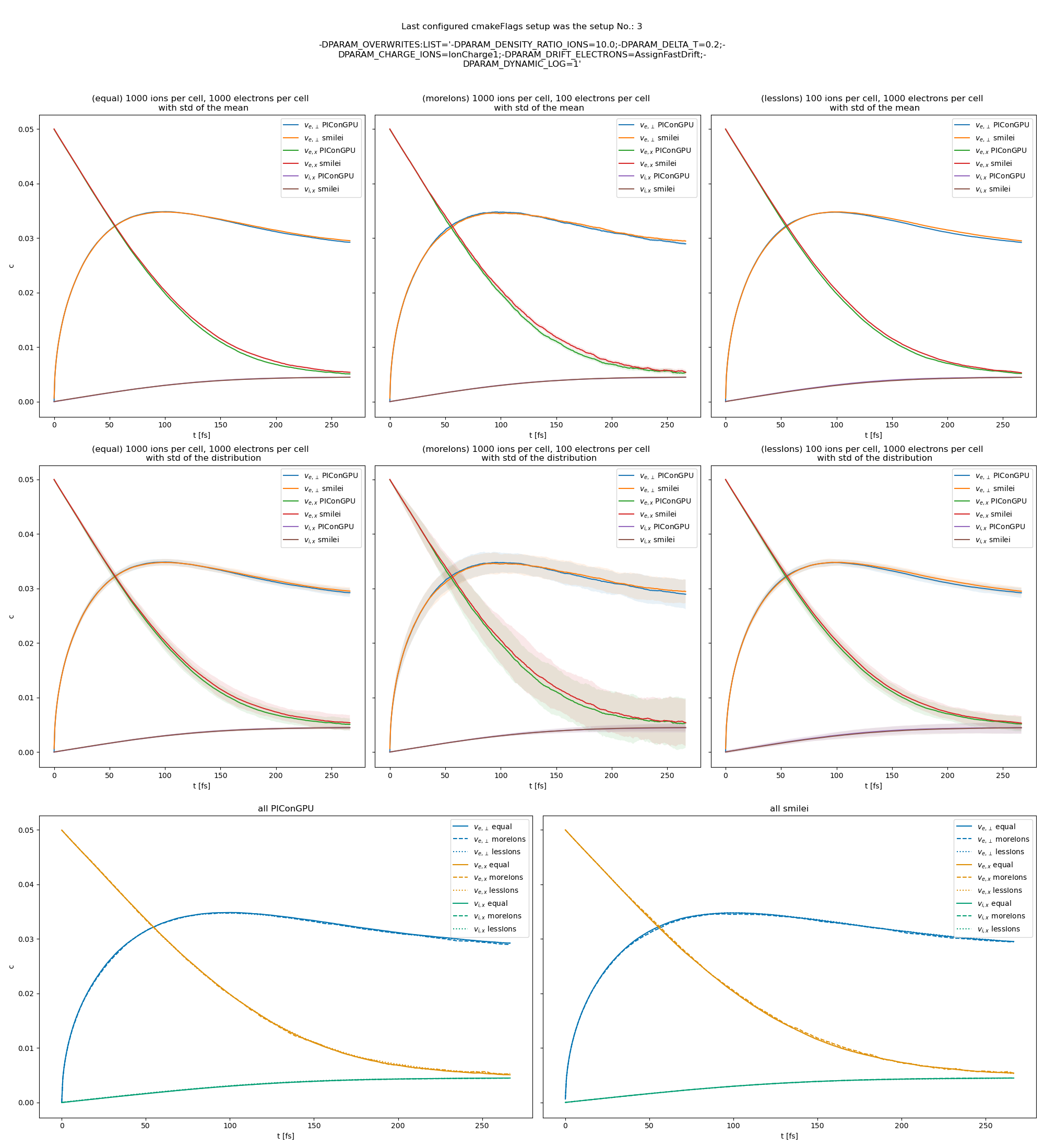
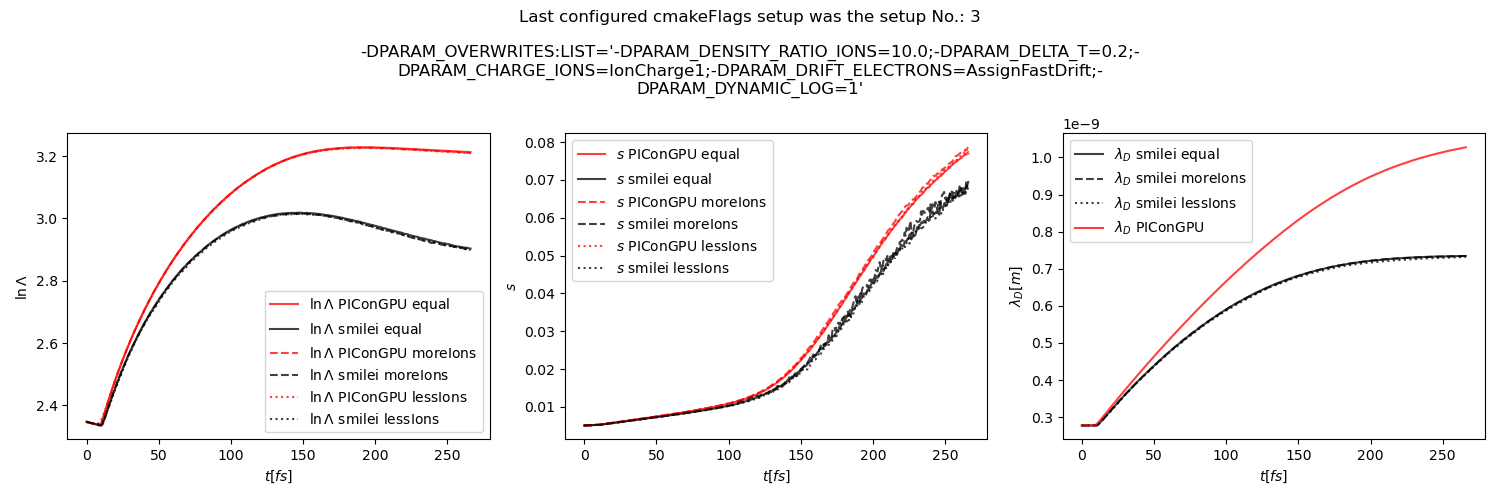 or this thermalization test:
or this thermalization test:
thermalization example set-up
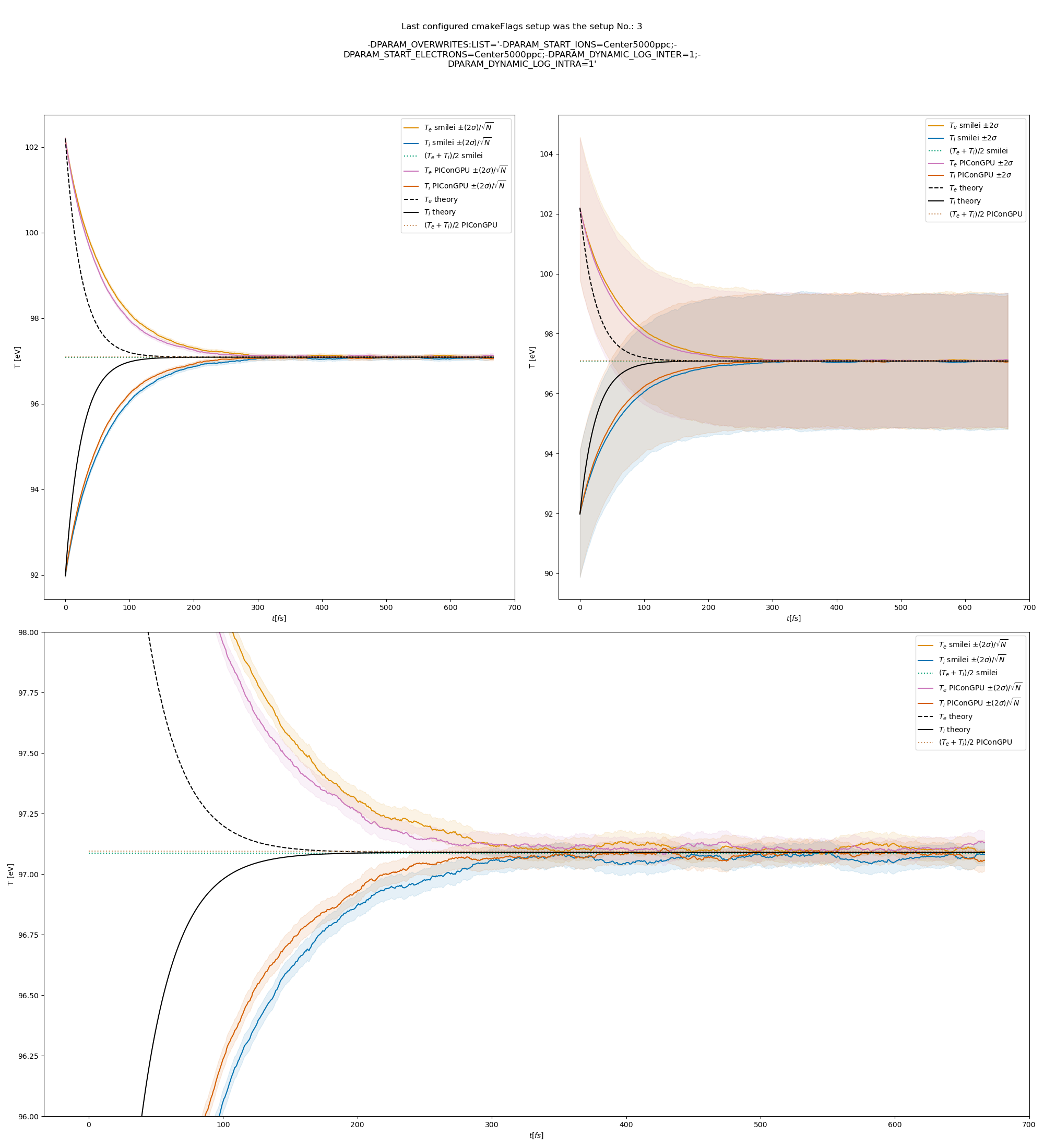
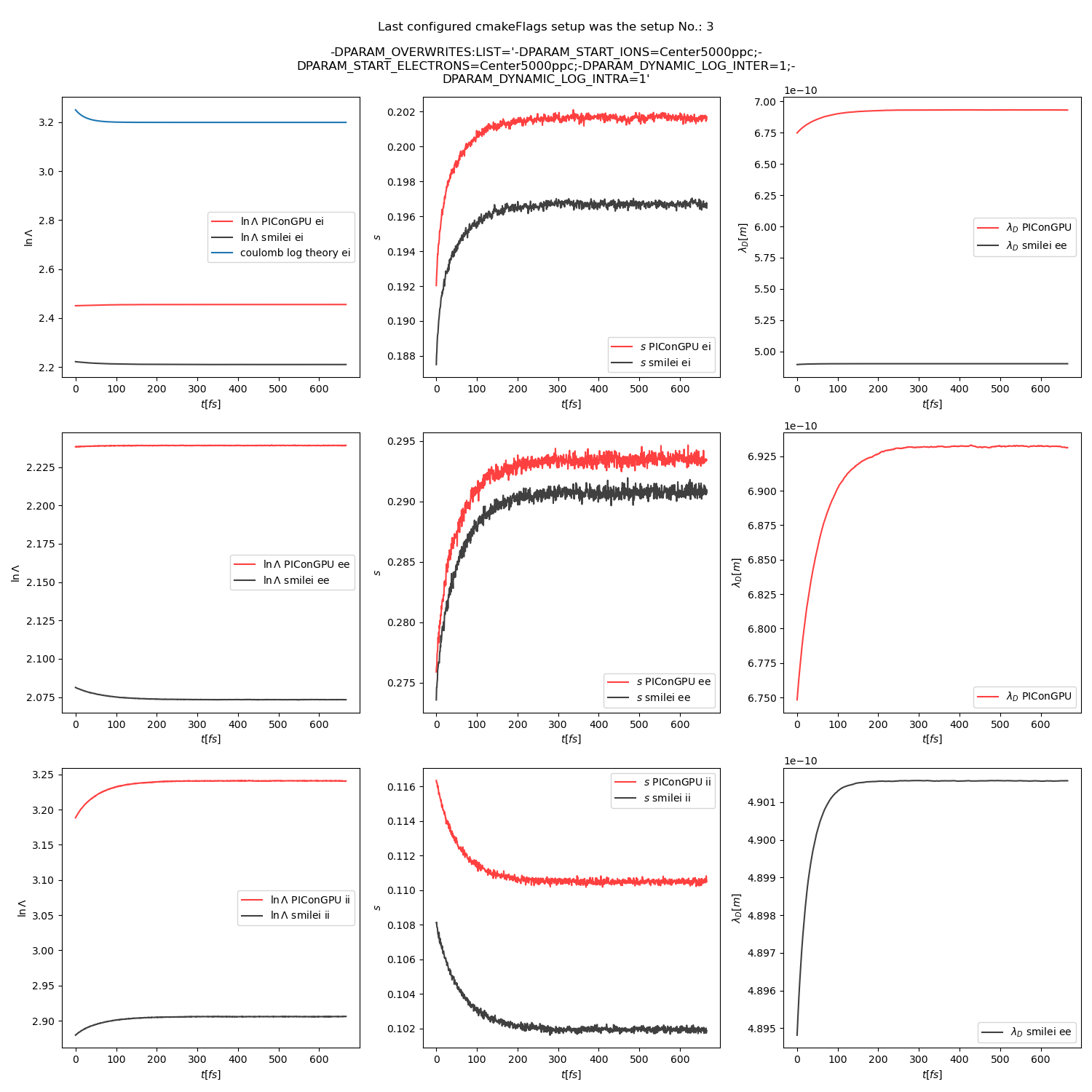
All the other combinations of particle weighting ratios, and coulomb logarithms are included in the updated docs.
The CI is not happy with your formation.
@psychocoderHPC as discussed offline, we should still try to investigate the difference in Debye length calculation. In the thermalization test the contributions from electrons and ions are similar, while in the beam relaxation test the ion contribution gets bigger overtime while ions gain energy. So, I think that the problem could be in ion contribution, but I don't know why yet.
@psychocoderHPC: fixed the missing absolute value, fixed the CI errors, updated the documentation with the new plots
@pordyna Could it be that you missed updating the plots in the first post of this PR? We should update those too to reflect the current state of the PR.
@psychocoderHPC I updated the first post
@psychocoderHPC If the CI passes, this PR is ready for review. I will rerun all tests once we are done with the review / introducing new changes.
@pordyna Why is in TH_debug_7.png the right plot for PIConGPU and smile differently?
CI has passed, I will rerun the tests
@psychocoderHPC tests results didn't change