MRI-to-CT-DCNN-TensorFlow
 MRI-to-CT-DCNN-TensorFlow copied to clipboard
MRI-to-CT-DCNN-TensorFlow copied to clipboard
This repository is the implementations of the paper "MR-based Synthetic CT Generation using Deep Convolutional Neural Network Method," Medical Physics 2017.
MRI-to-CT-DCNN-TensorFlow
This repository is an implementation of "MR‐based synthetic CT generation using a deep convolutional neural network method." Medical physics 2017 by Xiao Han.

Requirements
- tensorflow 1.13.1
- numpy 1.15.2
- opencv 4.1.0.25
- matplotlib 2.2.3
- pickleshare 0.7.4
- simpleitk 1.2.0
- scipy 1.1.0
MR-Based Synthetic CT Generation Results
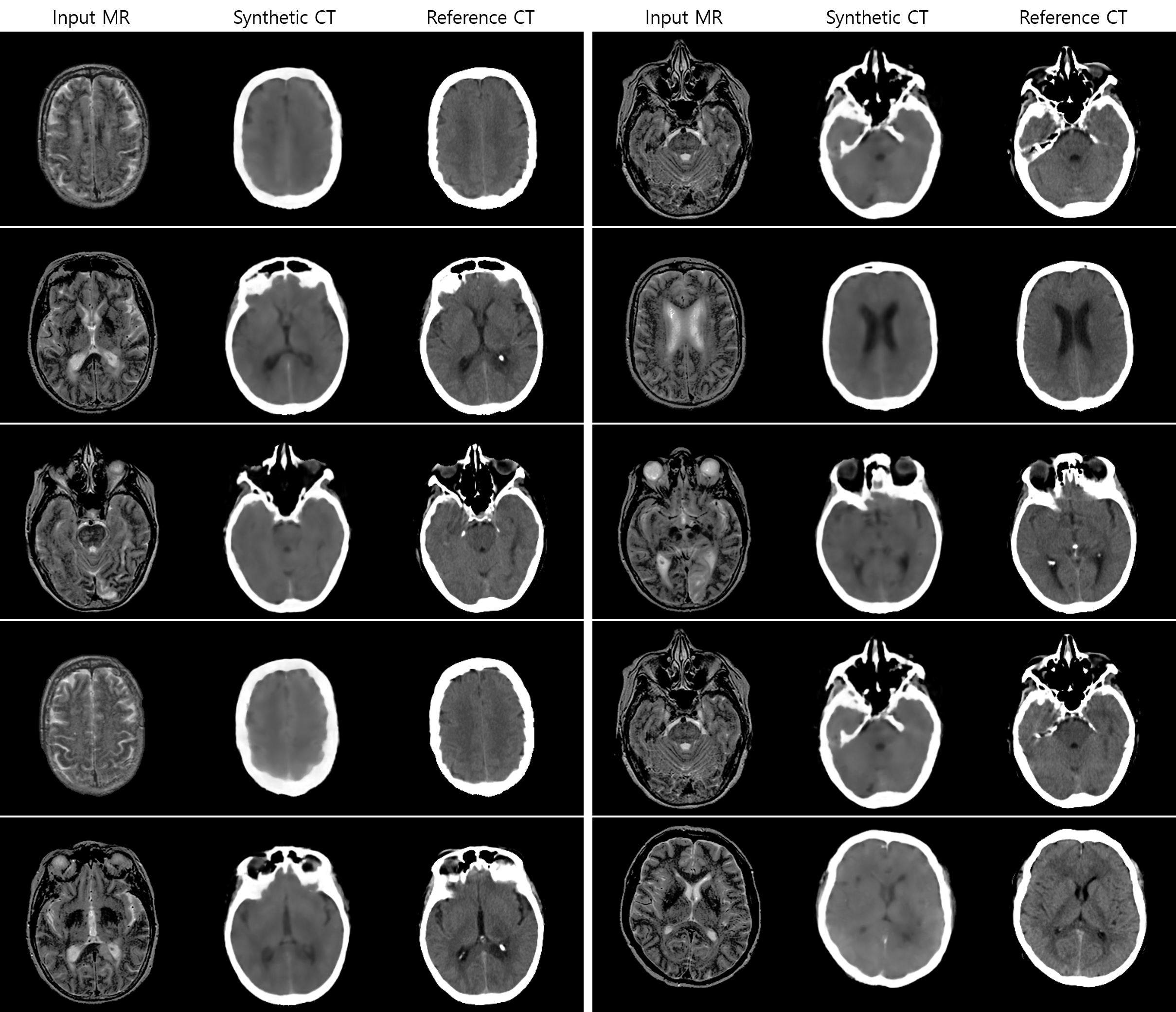
Implementations
- Six cross-validations
- Data preprocessing methods: N4 bias correction, histogram matching, and mask generation
- U-Net for CNN model
- Encoder of the U-Net is initialized by pretrained VGG16 weights
- Best model is saved based on the MAE evaluation in validation data
- Tensorboard visualization
- MAE, ME, MSE, and PCC metrics in test data
Documentation
Dataset
Download our toy dataset from here. This toy dataset just includes 367 paired images. We randomly divide data into training, validation, and test.
Directory Hierarchy
.
│ MRI-to-CT-DCNN-TensorFlow
│ ├── src
│ │ ├── dataset.py
│ │ ├── get_mask.py
│ │ ├── histogram_matching.py
│ │ ├── main.py
│ │ ├── model.py
│ │ ├── n4itk.py
│ │ ├── preprocessing.py
│ │ ├── solver.py
│ │ └── utils.py
│ Data
│ └── brain01
│ │ └── raw
│ Models_zoo
│ └── caffe_layers_value.pickle
Download the pretrained VGG16 weights from here (password:3ouy).
Data Preprocessing
Use preprocessing.py to rectify N4 bias correction, histogram matching, and head mask generation. Example usage:
python preprocessing.py --delay=1 --is_save
-
data: dataset path for preprocessing, default:../../Data/brain01/raw -
temp_id: template image id for histogram matching, default:2 -
size: 'image width and height (width == height), default:256 -
delay: interval time when showing image, default:1 -
is_save: save processed image or not, default:False
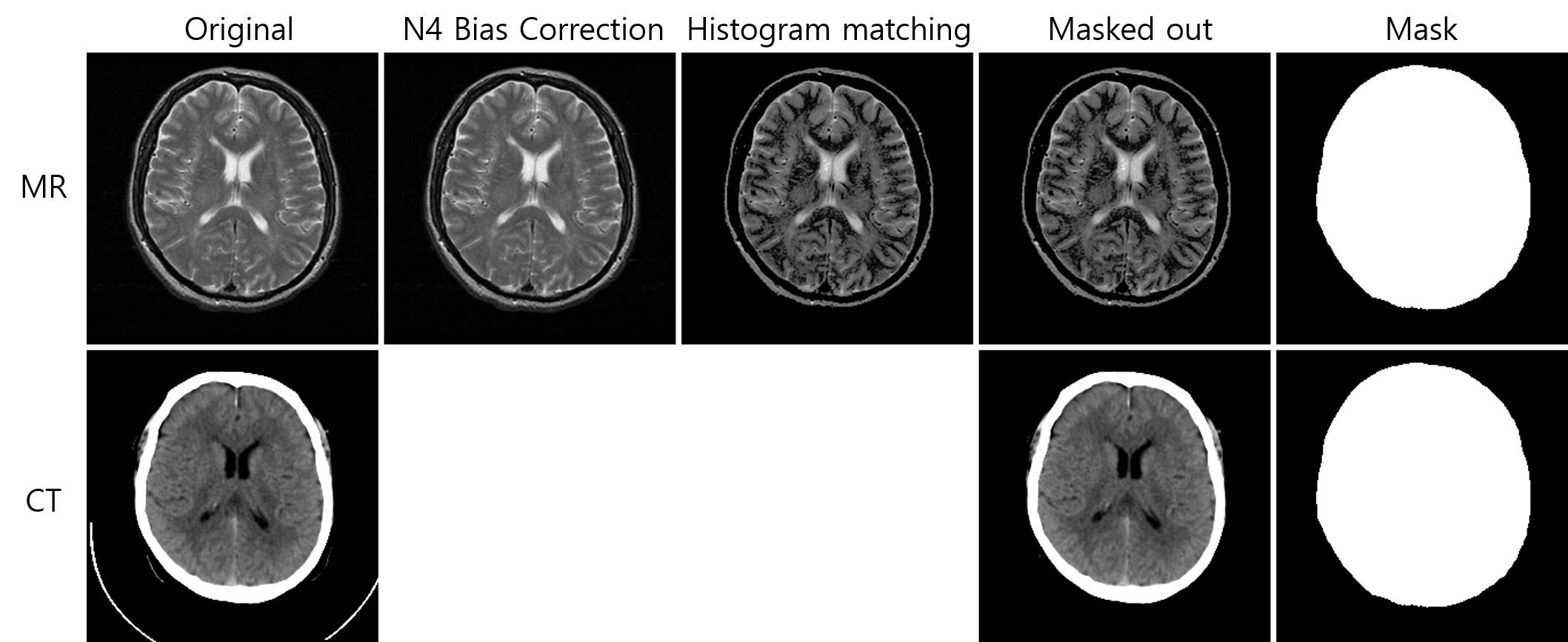


- N4 bias field correction using N4ITK
- Histogram matching using Dynamic histogram warping algorithm
- Binary head mask using Otsu auto-thresholding
Training DCNN
Use main.py to train a DCNN model. Example usage:
python main.py --is_train
-
gpu_index: gpu index if you have multiple gpus, default:0 -
is_train: training or test mode, default:False (test mode) -
batch_size: batch size for one iteration, default:8 -
dataset: dataset name, default:brain01 -
learning_rate: learning rate, default:2e-4 -
epoch: number of epochs, default:600 -
print_freq: print frequency for loss information, default:100 -
load_model: folder of saved model that you wish to continue training, (e.g. 20190411-2217), default:None
Test DCNN
Use main.py to test the DCNN model. Example usage:
python main.py --load_model=folder/you/wish/to/test/e.g./20190411-2217
please refer to the above arguments.
Tensorboard Visualization
Evaluation of the MAE, ME, MSE, and PCC in validation data during training process. Different color represents different model in six cross-validations.

Total loss, data loss and regularization term in each iteration.

Test Evaluation
MAE, ME, MSE, and PCC for six models and average performance.

Citation
@misc{chengbinjin2019DCNN,
author = {Cheng-Bin Jin},
title = {MRI-to-CT-DCNN-Tensorflow},
year = {2019},
howpublished = {\url{https://github.com/ChengBinJin/MRI-to-CT-DCNN-TensorFlow},
note = {commit xxxxxxx}
}
References
- Han, Xiao. "MR‐based synthetic CT generation using a deep convolutional neural network method." Medical physics 44.4 (2017): 1408-1419.
- Tustison, Nicholas J., et al. "N4ITK: improved N3 bias correction." IEEE transactions on medical imaging 29.6 (2010): 1310.
- Cox, Ingemar J., Sébastien Roy, and Sunita L. Hingorani. "Dynamic histogram warping of image pairs for constant image brightness." Proceedings., International Conference on Image Processing. Vol. 2. IEEE, 1995.
- Otsu, Nobuyuki. "A threshold selection method from gray-level histograms." IEEE transactions on systems, man, and cybernetics 9.1 (1979): 62-66.
License
Copyright (c) 2018 Cheng-Bin Jin. Contact me for commercial use (or rather any use that is not academic research) (email: [email protected]). Free for research use, as long as proper attribution is given and this copyright notice is retained.