bug in 'conditional_after' implementation
Bug: if you try putting the same timestamp in the conditional_after as in times, i.e.
t=10
cph.predict_survival_function(df, times=t, conditional_after=t)
The expected value, by the definition of 'conditional probability', is 1. However, you'd notice result from calling the function above isn't 1.
Calculating the conditional probability by hand (dividing two survival rates), the results don't match the function's result either (i.e. conditional_s_calc!=conditional_s_func).
t=10
s1 = cph.predict_survival_function(df, times=t).values
s2 = cph.predict_survival_function(df, times=t-1).values
conditional_s_calc = s1/s2
conditional_s_func = cph.predict_survival_function(df, times=t, conditional_after=t-1)
The bug is, In the source code, this line, the two coefficients are flipped, and the np.clip function is not necessary (can be removed once flipped IMO).
After flipping the order of partial hazards, it should work as expected.
Hi @louisazhou, this looks interesting, but I'm worried that this is a documentation problem instead. When using conditional_after, the timeline is moved up to start at the value of conditional_after. See LOC here.
IIRC, the reason we do this is because there would be a redundant sequence of 1s for all times before conditional_after.
With this in mind, do you still think there is a bug?
Hi @CamDavidsonPilon, I am not sure if I understand correctly. I still think the problem is the order of the two coefficients.
Let me elaborate my thought process. The equation below calculates the 'conditional probability' of P(t2|t1), given the cumulative hazard at t2 and t1, i.e. H(t2) and H(t1).
Note: if the equation isn't shown in dark mode, you might need to click the top empty space and open it in a new window to read
The last bit says that the conditional probability is equal to the difference of cumulative hazards at t1 and t2. t1 is the timestamp of the first event (i.e., the time we use in the 'conditional after'). Therefore, the baseline hazard returned in the if conditional_after is not None: clause should be
c_0 = (c_0_conditional_after - c_0).T
instead of
c_0 = np.clip((c_0 - c_0_conditional_after).T, 0, np.inf)
Because c_0_conditional_after >= c_0 naturally (the baseline survival at later time is by definition smaller or equal to that at a previous timestamp), there is not need to use a clip function to clip the results to a positive range.
Could you enlighten me on what I might have gone wrong or whether there is something else about the start time that I got mixed up?
We can rewrite your equation as:
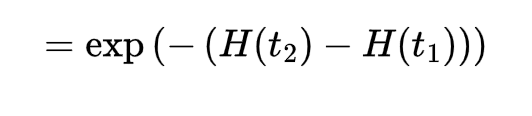
So really the conditional cumulative hazard rate is H(t2 | t_1) = H(t_2) - H(t_1). Do you agree?
By definition, H(t) is an non-decreasing function, so if t_2 > t_1, then H(t_2) >= H(t_1).
In the LOC here:
times_to_evaluate_at = np.tile(times_, (n, 1)) + conditional_after
c_0 = utils.interpolate_at_times(self.baseline_cumulative_hazard_, times_to_evaluate_at)
c_0_conditional_after = utils.interpolate_at_times(self.baseline_cumulative_hazard_, conditional_after)
The values in c_0 are greater than c_0_conditional_after, so the final difference in
c_0 = np.clip((c_0 - c_0_conditional_after).T, 0, np.inf)
should be non-negative. I clip to force this, too.
If you disagree, please continue asking questions! This is useful for me, and there may be a subtle bug here.
@CamDavidsonPilon Thank you! I now see why you said it is a 'documentation problem': In order to evaluate the conditional probability of P_1/P_2, the input to this function should be
cph.predict_survival_function(df, times=t2-t1, conditional_after=t1)
because the start_time is moved but the returned index from the function is still the original timeline index (line 2382, index=times_).
This explains a lot. The provided example in the documentation did not specify the times= argument so I mis-interpreted the return value in the first place, and carried this misunderstanding in reading the code.
Thanks a lot for developing this package and for your fast response in this community!
I'm happy it's resolved, but I'm going to reopen this issue as a reminder to make the docs more clear
I'm happy it's resolved, but I'm going to reopen this issue as a reminder to make the docs more clear
please let me know what changes you want to see in the documentation, it is clear in my idea
@Unessam thanks for volunteering - I think an example would be sufficient that demonstrates the interaction between times and conditional_after
@CamDavidsonPilon thanks for the reply, do you think the below example can clarify the interaction?
` from lifelines import CoxPHFitter from lifelines.datasets import load_rossi
rossi = load_rossi()
cph = CoxPHFitter() cph.fit(rossi, duration_col='week', event_col='arrest')
filter down to just censored subjects to predict remaining survival
censored_subjects = rossi.loc[~rossi['arrest'].astype(bool)] censored_subjects_last_obs = censored_subjects['week']
predict new survival function
times = [10, 20, 30, 40, 50] cph.predict_survival_function(censored_subjects, times=times, conditional_after=censored_subjects_last_obs)
predict median remaining life
cph.predict_median(censored_subjects, conditional_after=censored_subjects_last_obs) `
hey guys, curious about the latest development of this.