Search
 Search copied to clipboard
Search copied to clipboard
Train and evaluate first RE models for `(GENE, is-in, BRAIN_REGION)`
Context
- In #606 we collected annotated sentences for the relation extraction (RE) task for the relation type
(GENE, is-in, BRAIN_REGION). - Effectively, this can be seen as an example of sentence classification.
- This Issue is now about implementing and evaluating different RE model.
Actions
- [ ] What is the percentage of sentences that the human expert discarded due to wrong NER annotations? Note that
- [ ] Out of the annotated samples, what is the percentage of samples that are labelled as "not is-in"?
- [ ] (Given the previous point:) What is the performance (precision, recall, f1-score) of a naïve RE model that predicts
is-inanytime there's a co-mention of aGENEand aBRAIN_REGIONin the same unit of text? - [ ] What is the performance of a pre-trained NLI model that takes as
premisethe paragraph and ashypothesisa sentence like[gene] is located in [brain region]? - [ ] What is the performance of a pre-trained binary-QA model that takes as
contextthe paragraph and ashypothesisa sentence likeIs [gene] located in [brain region]?? - [ ] What is the performance of a custom trained relation-extraction/sentence-classification model trained on our data? Note that unlike the previous pre-trained and naïve models, in this case we will need a 5-fold cross validation and compute corresponding mean and std for the various performance metrics
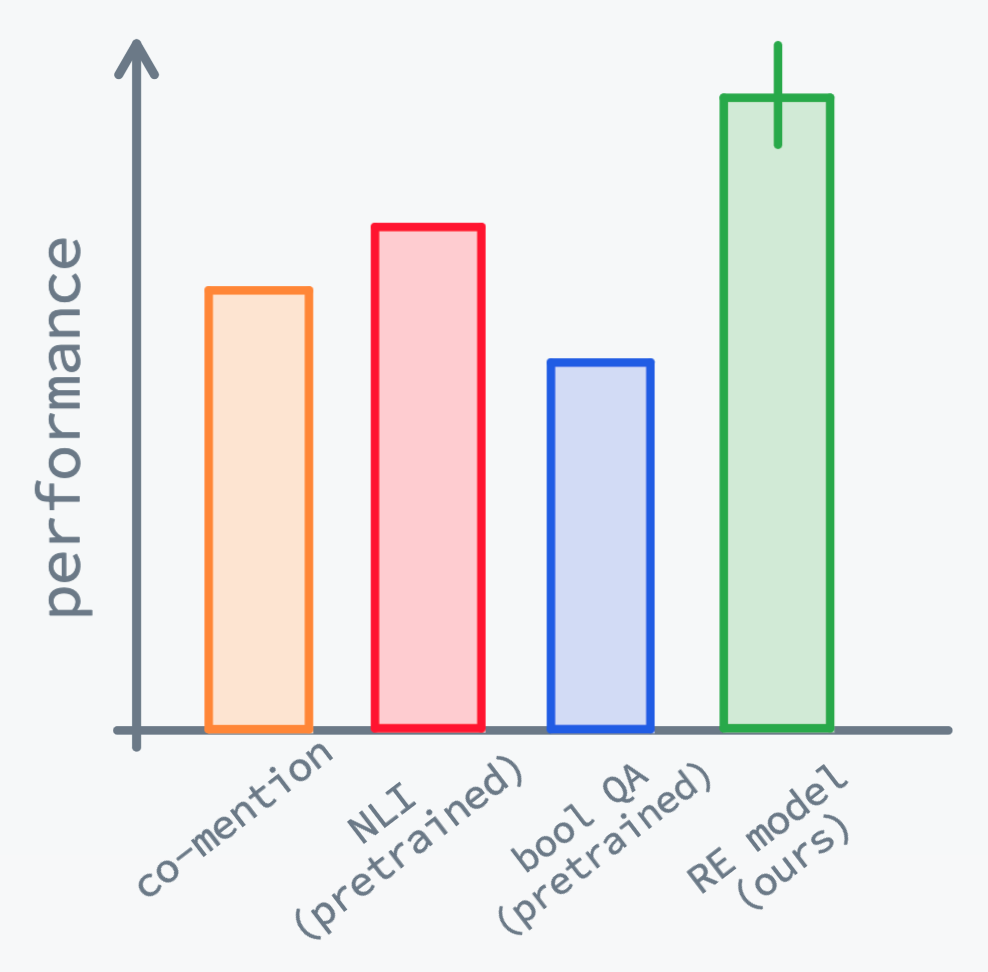
- [ ] Summarize all results in a histogram like the following:
Dependencies
- Before starting this Issue, #606 must be completed.
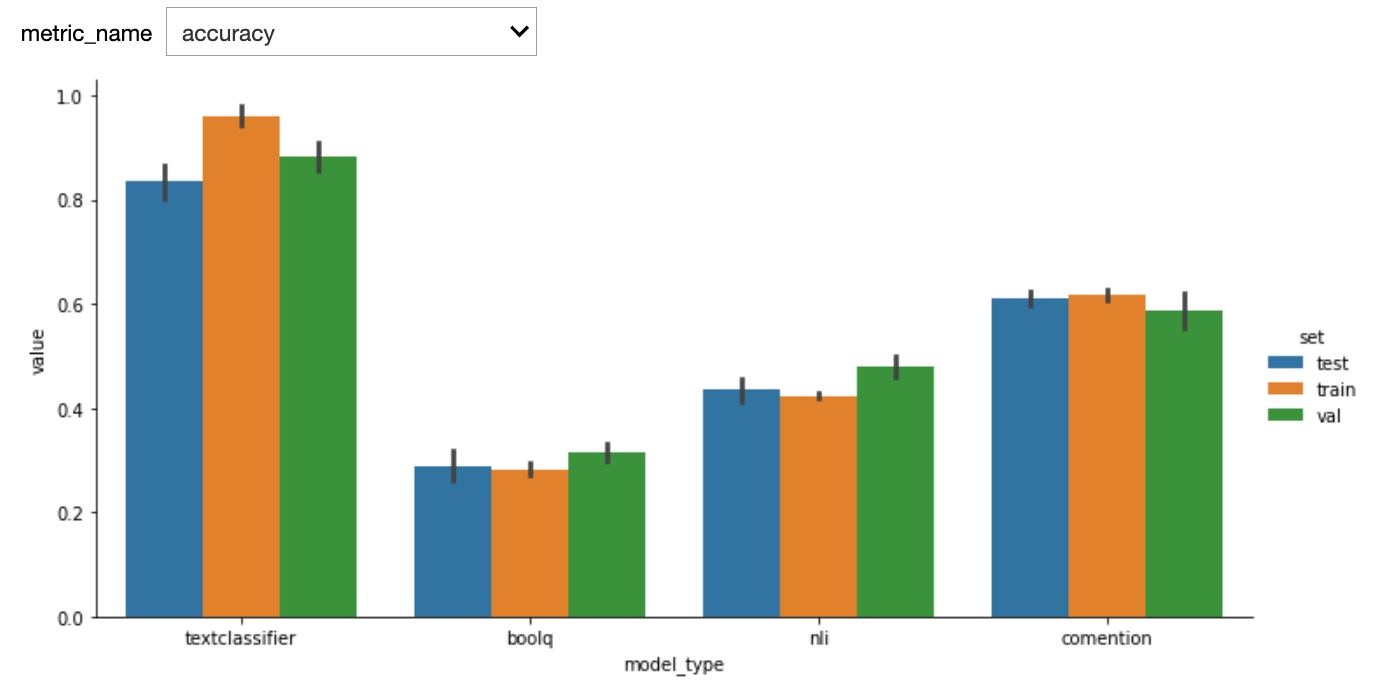
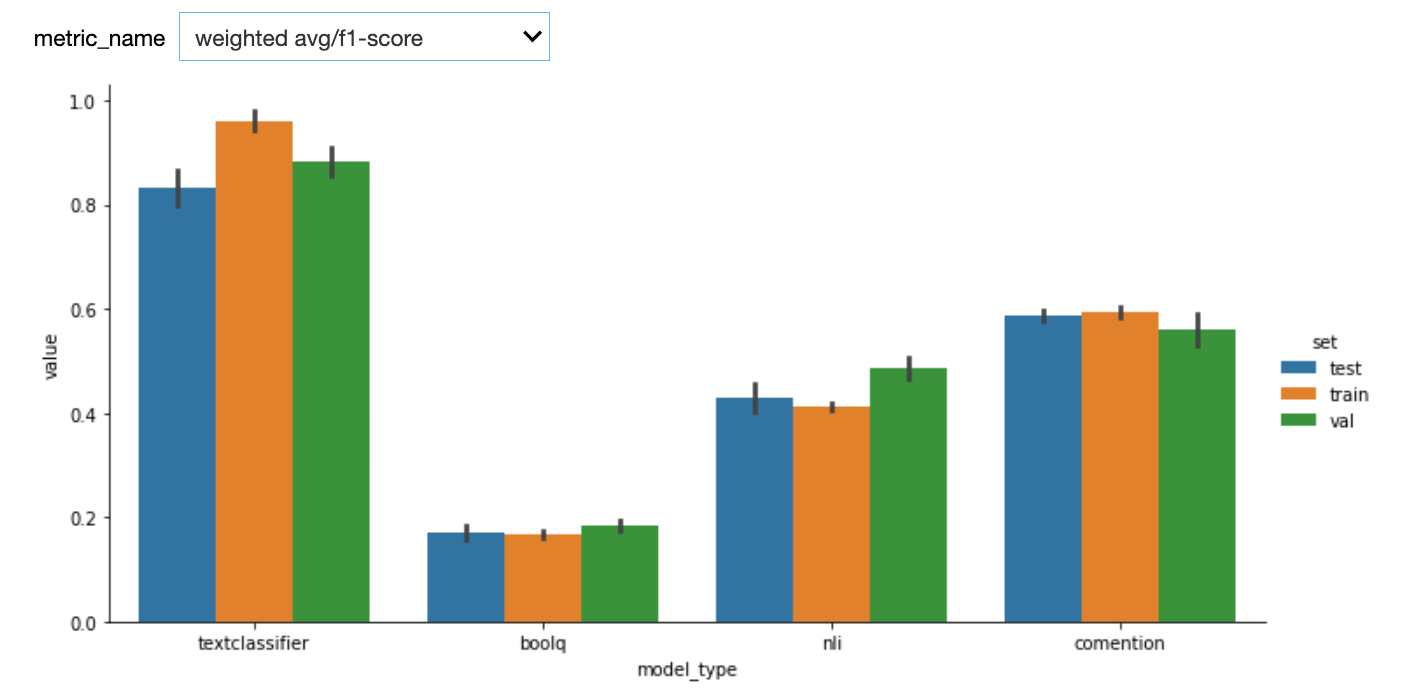
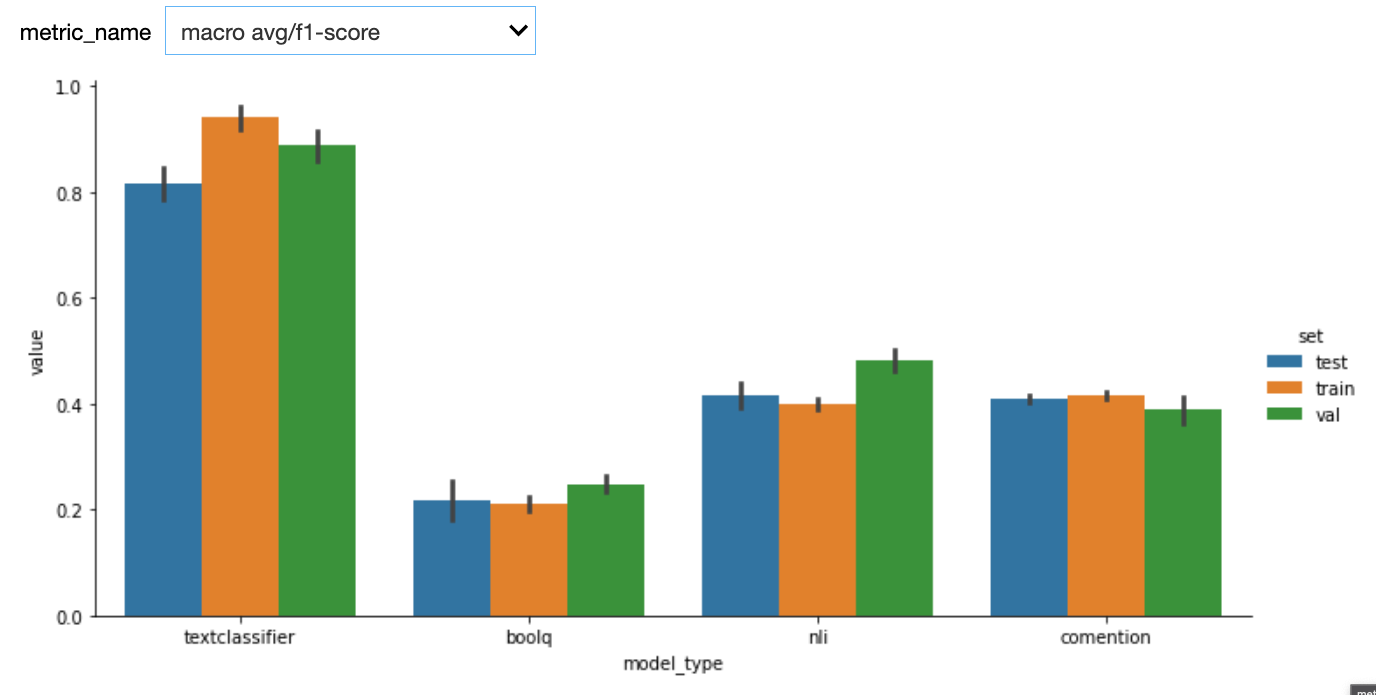
Here are the first results. We used StratifiedKFold with n_splits=5. One potential bias of our custom model (textclassifier) is that we used the the validations sets to do early stopping.
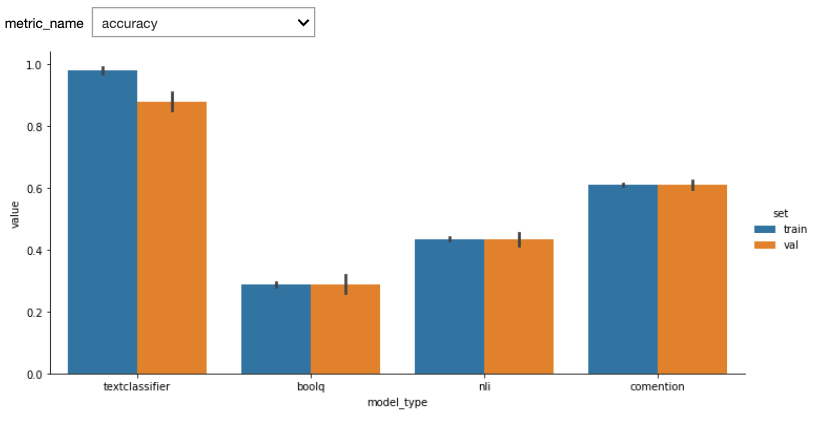
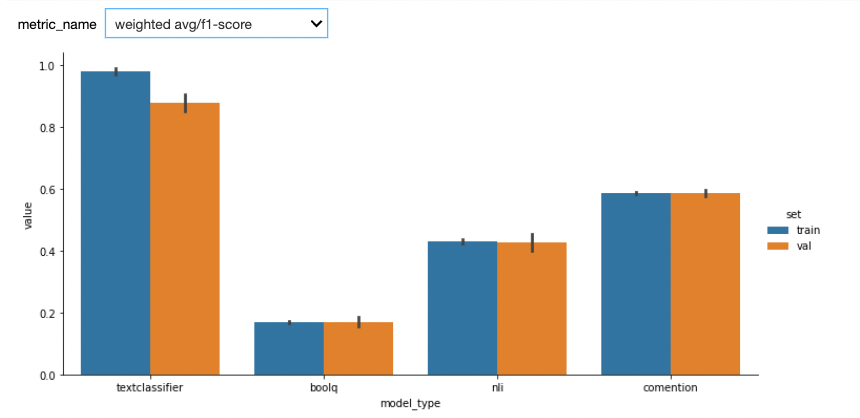
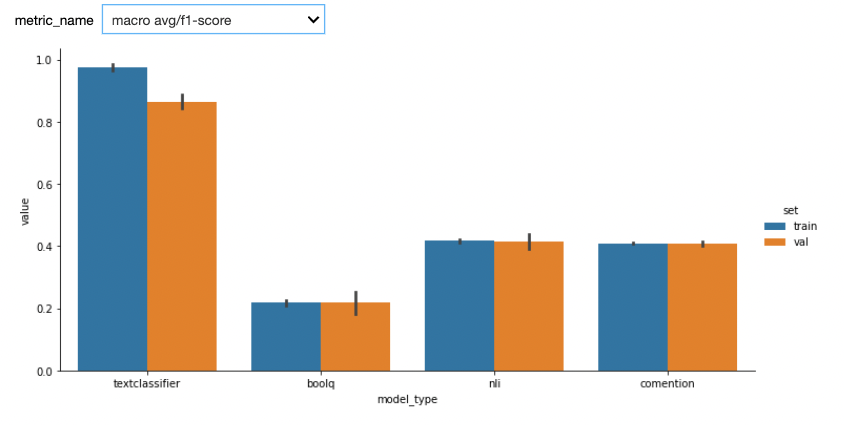
See below results after adding a test set