TCGAbiolinks
 TCGAbiolinks copied to clipboard
TCGAbiolinks copied to clipboard
Survival data inconsistencies
Dear all,
We used your useful package to download the TCGA-SKCM dataset. However, we have inconsistencies according to follow-up times (days_to_last_follow_up) and times until death (days_to_death):
- Sometimes, there was no follow-up times (probably missing data?)
- When the 2 follow-up times were provided, the time until death was always greater than follow-up time, but should the last follow-up not be the death (days_to_last_follow_up == days_to_death)?
- The vital status (« alive » or « dead ») of patients did not always fit with the 2 follow-up times. For example, 2 « alive » patients had a value for days_to_death, 2 « dead » patients had no value for days_to_death, and 7 « alive » patients had no value for days_to_last_follow_up.
- There are 3 negative values for follow-up times while it is not possible according to the variable definition. It should not be an indicator code for missing data because we have different negative values.
We contacted the TCGA support which had not these issues on their own database.
Do you have an idea what could be the origin of these issues?
The code we used to download the data:
library(TCGAbiolinks)
library(SummarizedExperiment)
query.exp <- GDCquery(
project = "TCGA-SKCM",
legacy = FALSE,
data.category = "Transcriptome Profiling",
data.type = "Gene Expression Quantification",
workflow.type = "HTSeq - Counts",
experimental.strategy = "RNA-Seq"
)
GDCdownload(query.exp)
skcm.exp <- GDCprepare(query = query.exp, save = TRUE, save.filename = "skcmExp.rda")
We also tried the procedure described here: https://rdrr.io/bioc/TCGAbiolinks/f/vignettes/clinical.Rmd
Many thanks in advance,
Best regards
Hello, I am checking the question. I'll answer them separate, because I am looking at the data and questions.
- Sometimes, there was no follow-up times (probably missing data?) Yes, no follow-up should means there is no data available. I'll check if the XML has data that the GDC does not have, but I don't think that should happen.
-There are 3 negative values for follow-up times while it is not possible according to the variable definition. It should not be an indicator code for missing data because we have different negative values.
Yes, indeed there are 3 negative values.
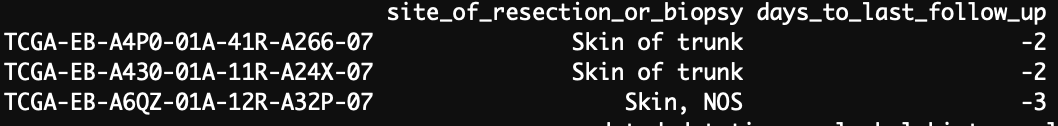
But they are also in GDC.
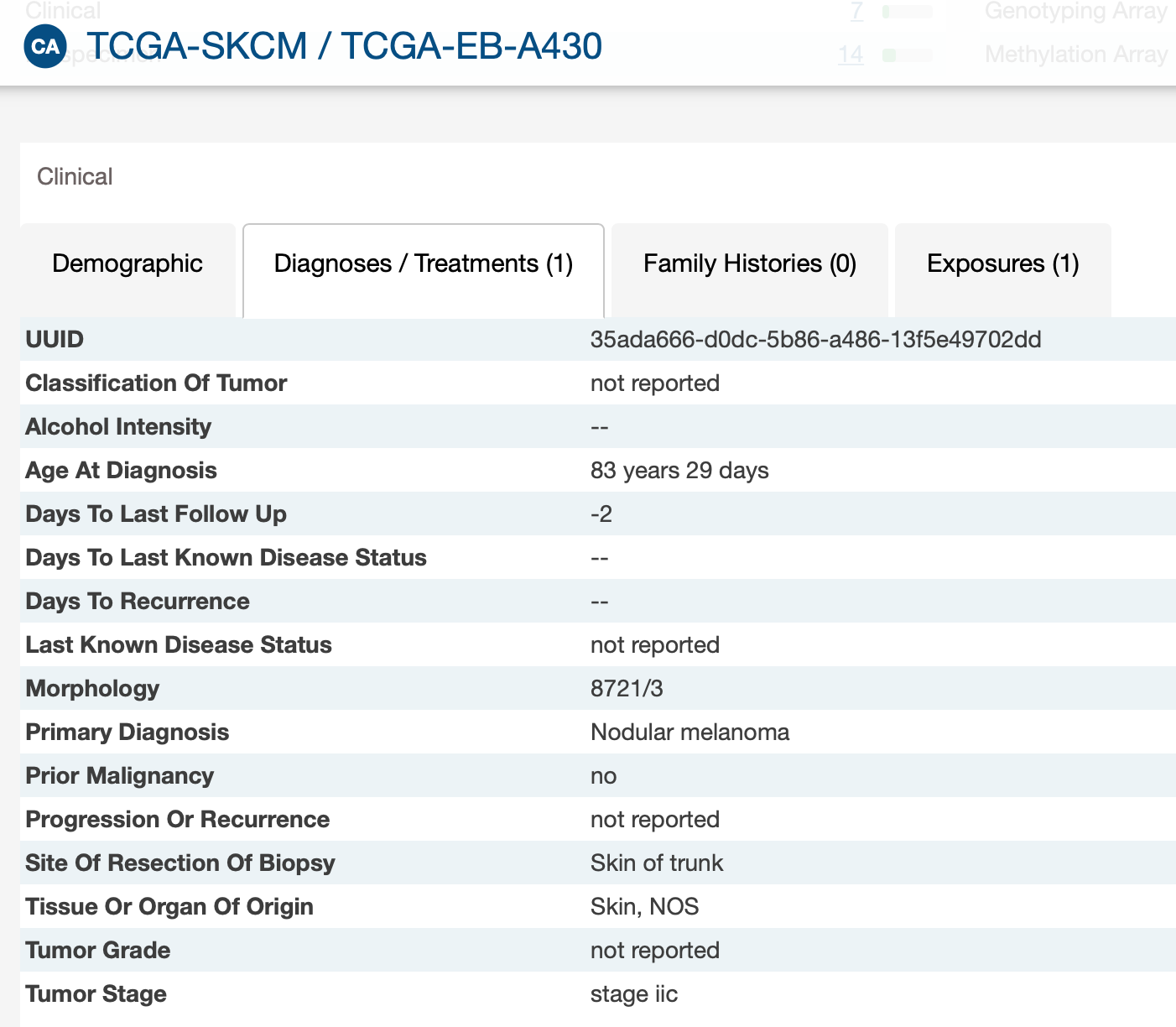
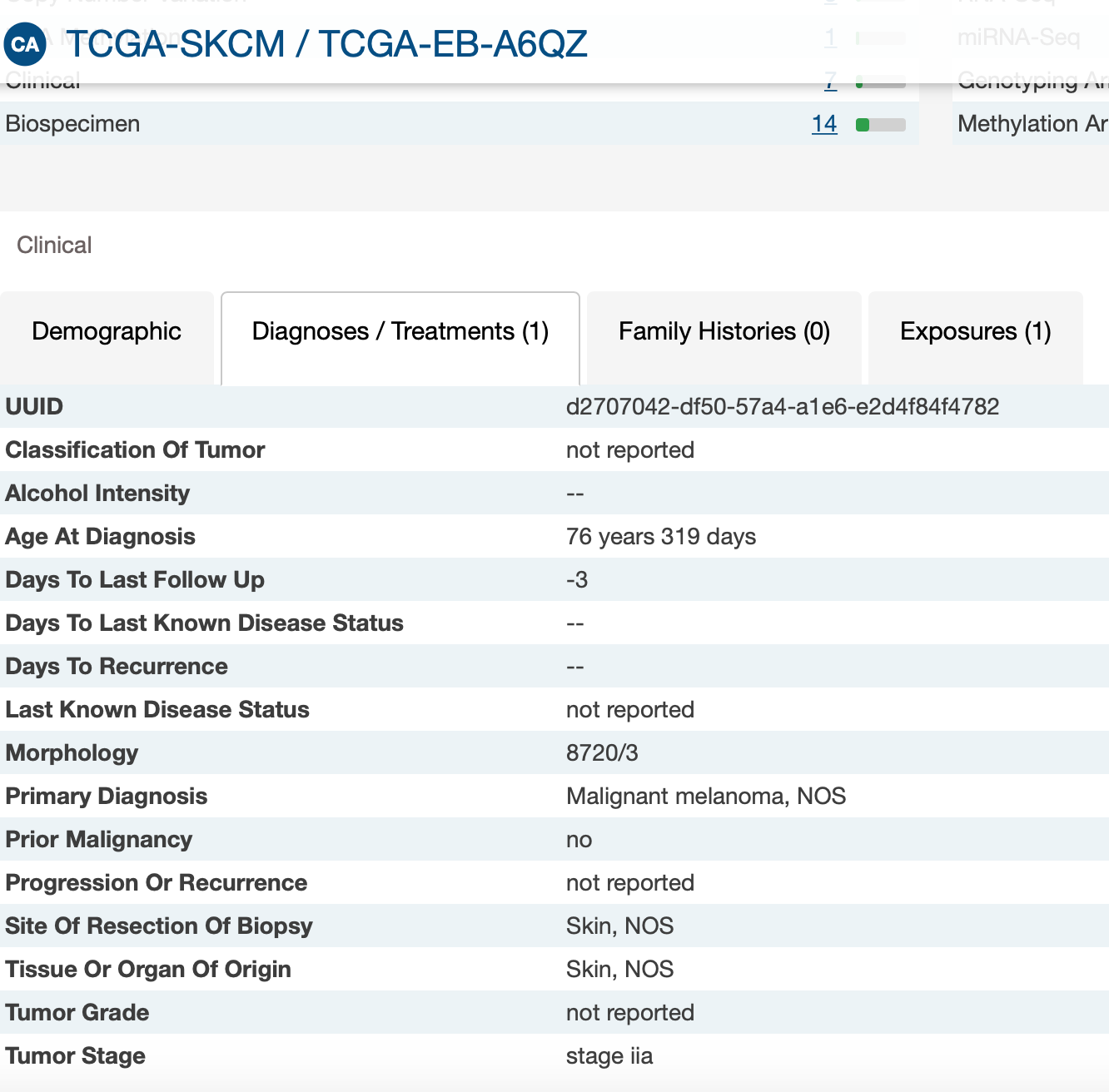
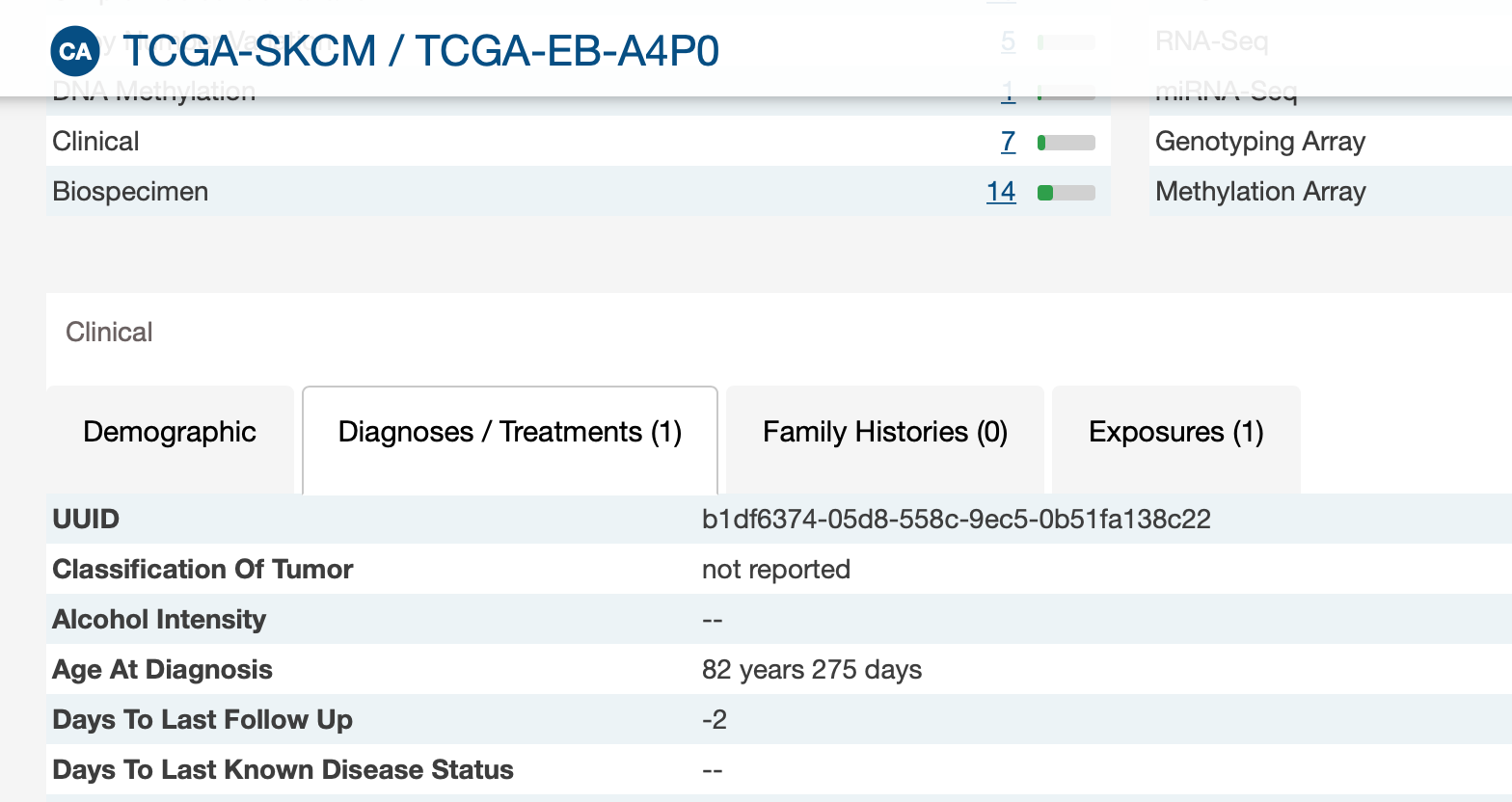

I am checking the XML, the samples with negative values, actually has follow-up data, but don't have the days to last follow up information (last_contact_days_to)

By "When the 2 follow-up times were provided, the time until death was always greater than follow-up time, but should the last follow-up not be the death (days_to_last_follow_up == days_to_death)?"
I understood the two follow-up times are days_to_death and days_to_last_follow_up. And you expect them to be the same. How many cases do you have that?
These are the XML cases with Dead and follow up.
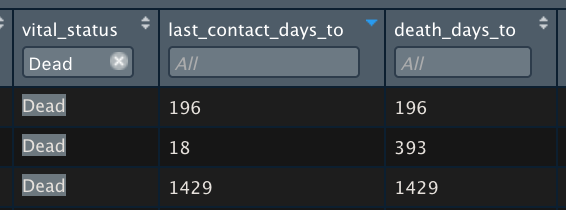
But I was checking the object data and we have 66 cases. So, my answer to the question is no. days_to_last_follow_up is not the same as days_to_death.
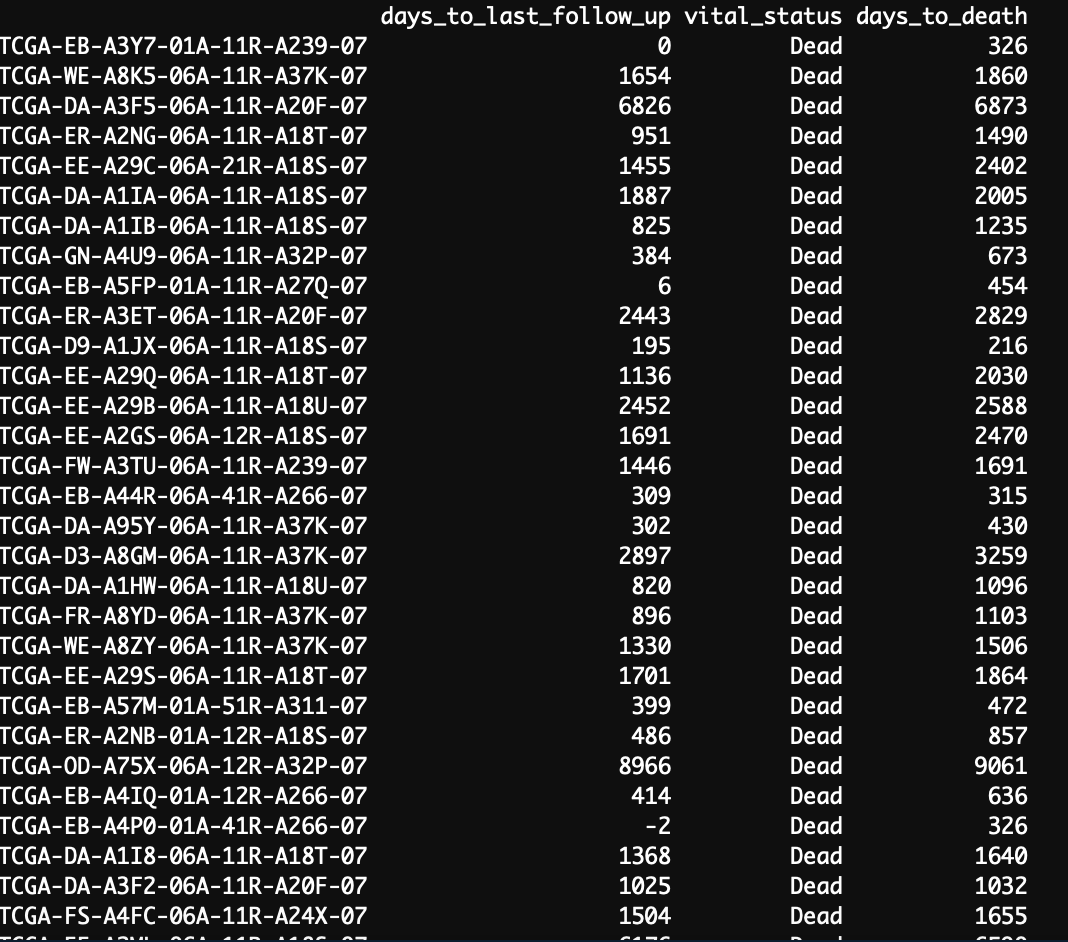
But I am still surprised with some values. I am not sure how they calculate the 6 days to follow up if that information is not in the XML files.

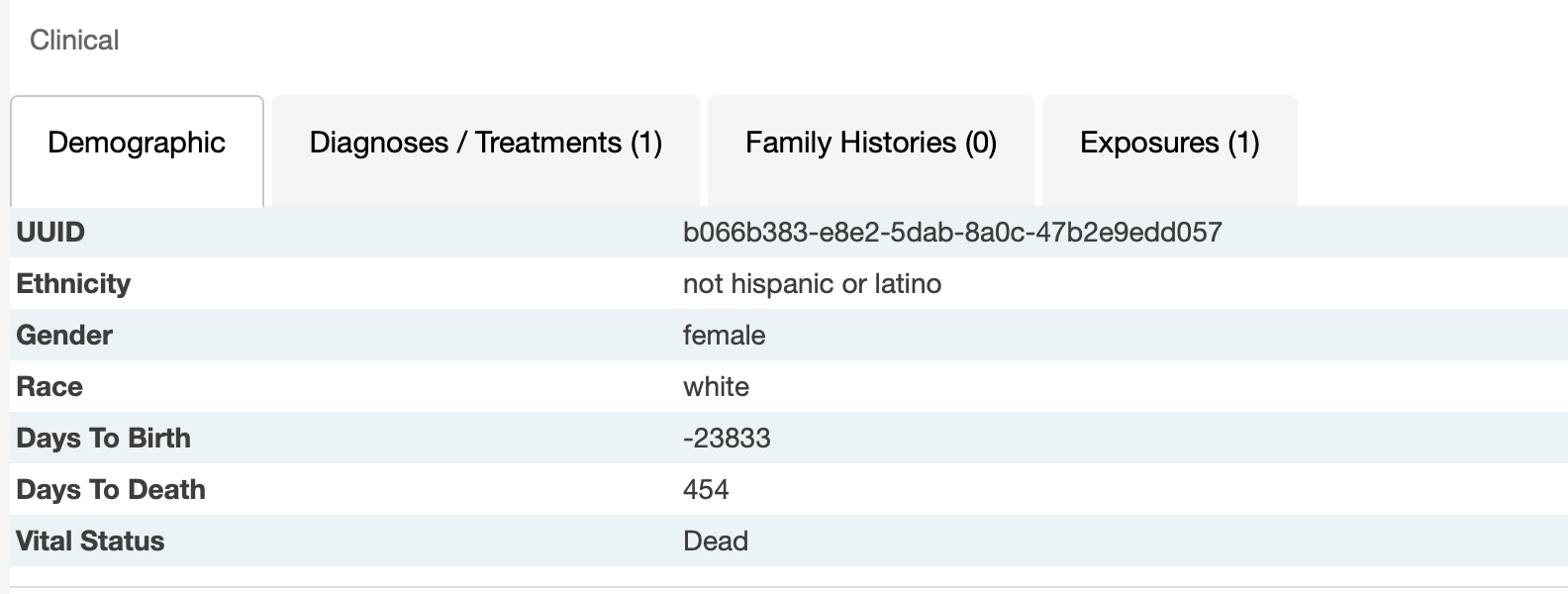
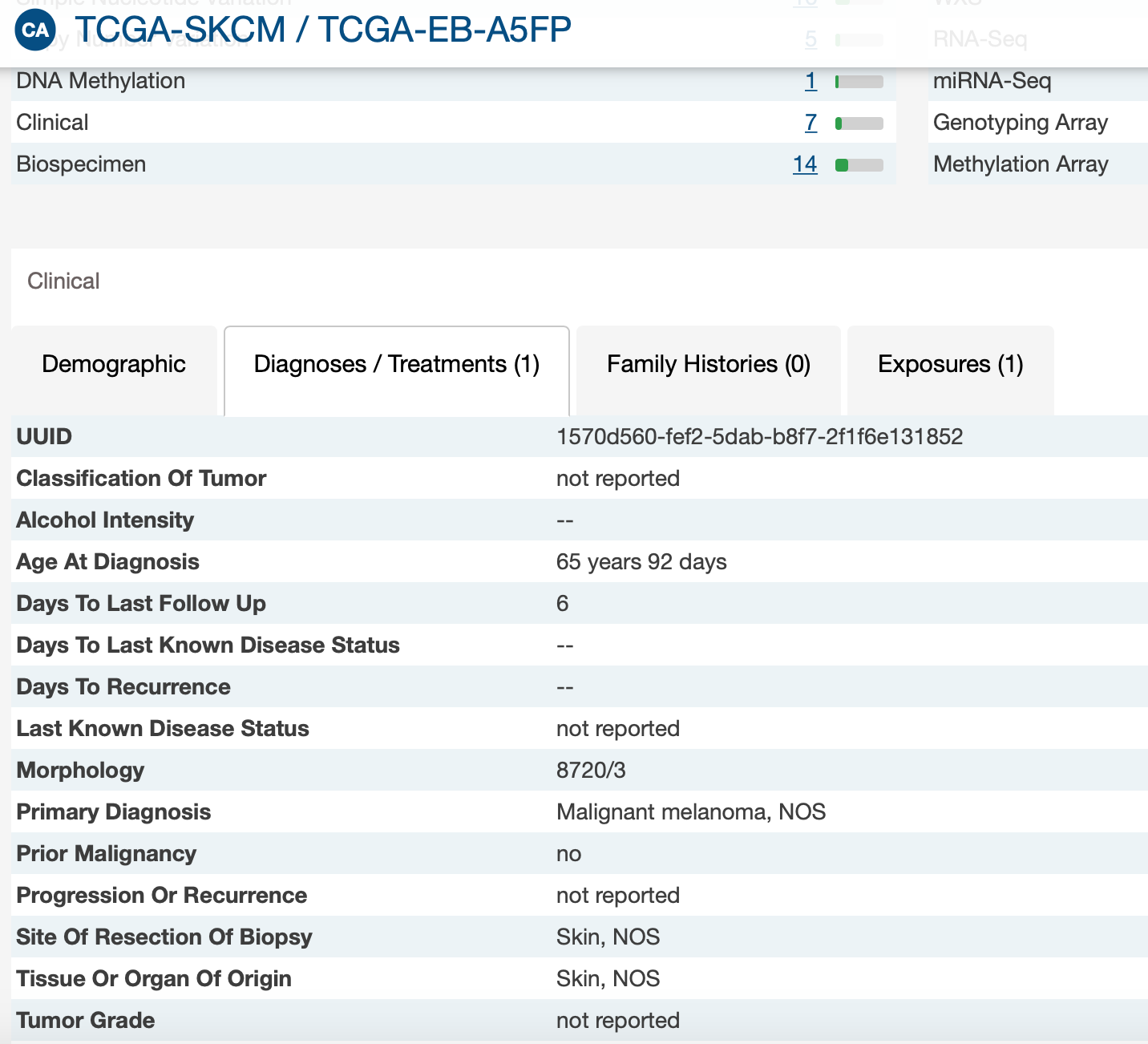
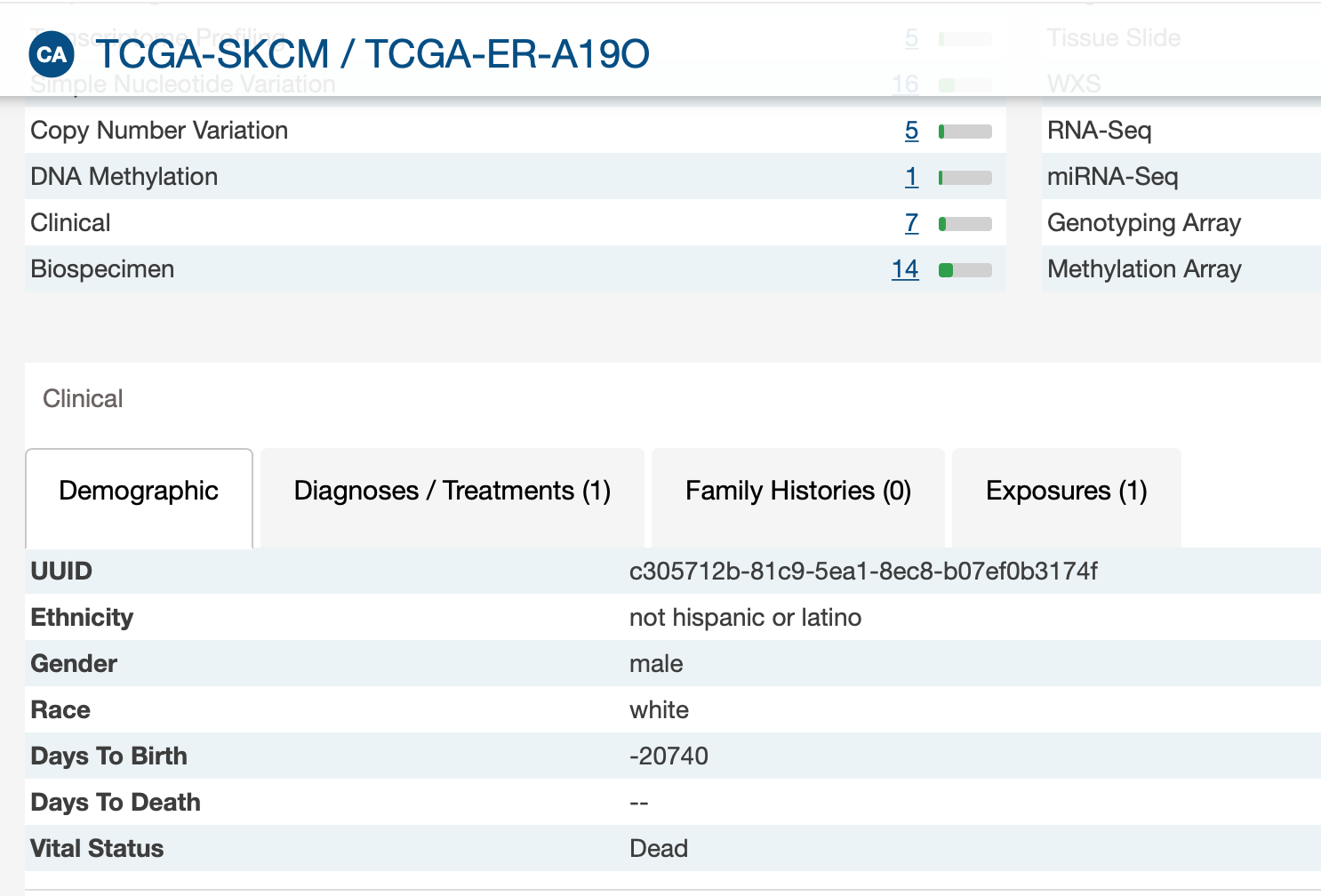
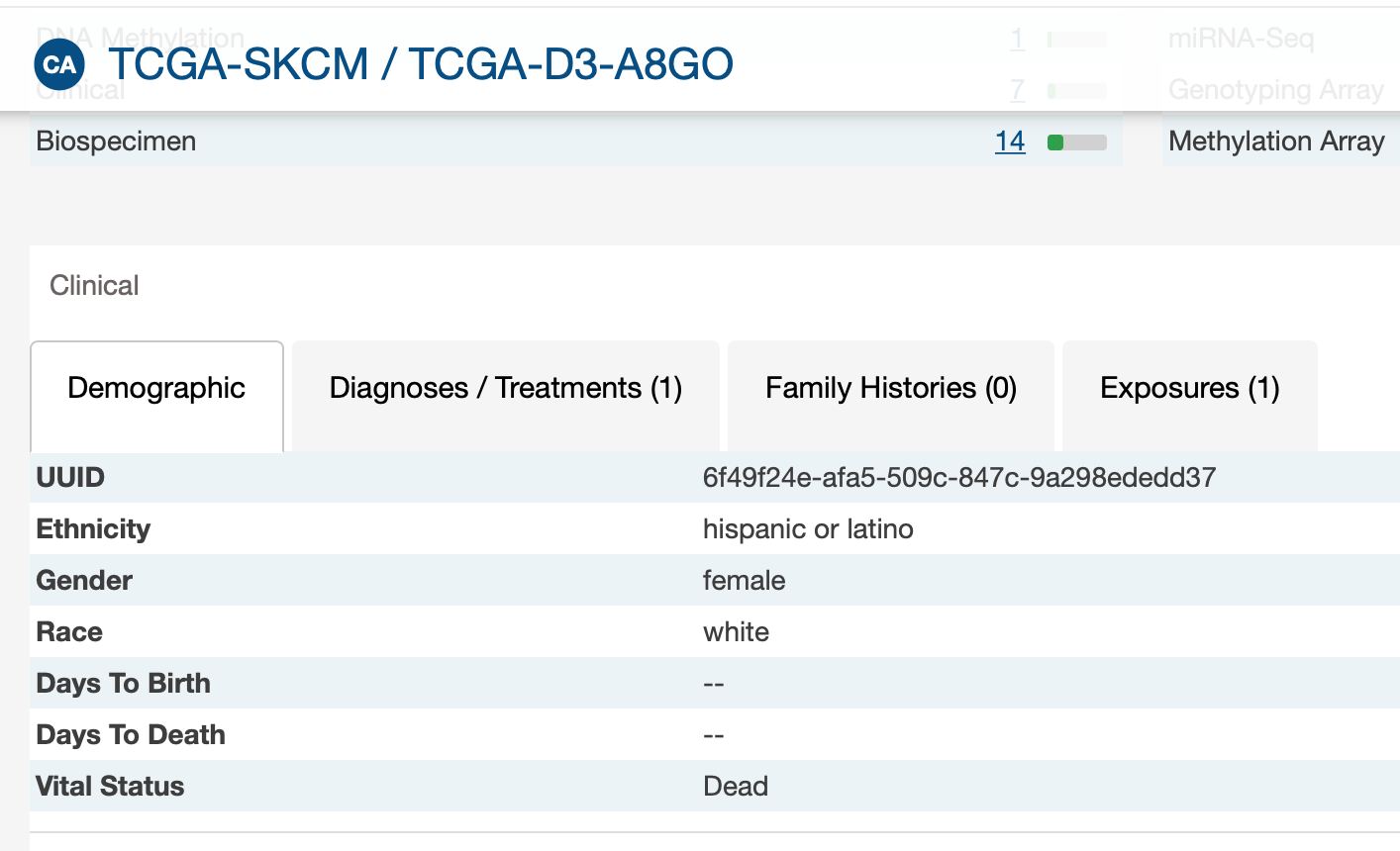
- The vital status (« alive » or « dead ») of patients did not always fit with the 2 follow-up times. For example, 2 « alive » patients had a value for days_to_death, 2 « dead » patients had no value for days_to_death, and 7 « alive » patients had no value for days_to_last_follow_up.
I don't have patient alive with days_to_death. And I believe there might be patients without days_to_last_follow_up.
But I have dead patients without days_to_death as GDC.
Here is the GDC answer.
Hello Tiago,
Thank you for contacting the GDC Help Desk.
I have talked to our Clinical Data Scientist and she said that these values can seem strange due to how TCGA and third parties handled patient data. For the negative follow up dates, this can be caused by the pathology report being made at a secondary institution that does the surgery, even though the patient was diagnosed at a local hospital. The larger institution/center eventually reports back the pathologic findings from the surgery, but they never receive follow-up information about the patient. Thus, the days to diagnosis would have been the date the pathology report was signed out, which is always day 0, but the last time the patient was seen relative to that date could have been days or weeks prior to that report being signed out.
Basically, we are aware that these negative dates exist, and they are caused by pathology report dates taking priority as day 0.
For the patients without days_to_death, this is the case that they knew the patient was deceased, but they were uncertain of the exact date. Thus, when filling out the clinical information they left this intentionally blank.
Please let me know if you have any further questions.
Best regards,
Sean Burke, Ph.D. Scientific Support Analyst
Many thanks for this complete answer ! We will deal with that.
Best regards
@tiagochst so shall we delete the negative values and 0 when calculating over survival? if do not delete it ,will it get a wrong conlusion thanks a lot?