pyart
 pyart copied to clipboard
pyart copied to clipboard
PhiDP unfolding issues using phase_proc_lp
pyart.correct.phase_proc_lp seems to have issues with IRIS RAW files. I am trying to use this function to recalculate KDP values, but as it can not correctly unfold the PhiDP values, it affects the recalculation of KDP as well. There are no issues in rays with no folding of PhiDP, but in rays where folding occurs it is problematic: it starts with a negative value and the highest value in these rays is 0 degrees.
The original PhiDP data with folding in some rays
Unfolded PhiDP using phase_proc_lp
Recalculated KDP has similar signature as a result:
Playing around with nowrap and overide_sys_phase parameters did not seem to help either. The first did not have any effect seemingly and the second one changed all the values, but the unwanted behaviour resisted (as was expected).
As a quick and dirty fix I can think of adding a check for each ray which reads the minimum negative PhiDP value (if present) and adds this to each bin in this ray.
To check the original file I uploaded it here (NB! the file has original PhiDP saved in "reverse", to fix it one should change the sign and add 180 to PhiDP original data, e.g. [orig_phidp*(-1)+180] and the KDP is negative) SUR160703101507.zip
Has anyone noticed something similar with their files? Do you have any suggestions why this is happening?
Thanks! Yes. The unwrap code needs a complete rework.. It also needs to take in a gate filter as an input.. It was written quickly to fix an issue.. However it should matter what file it comes from.. To Py-ART data is data..
I’ll leave it to the community for a little while to take a look before giving it a crack myself.. But we do know (not just for Py-ART but for our ARM work) this is something we need to fix.
Another thing you could try (that I have been meaning to) is to shoe-horn the data into the region based dealiaser.
-- Scott Collis
On Oct 31, 2017 at 8:36 AM, tanelv [email protected] wrote:
pyart.correct.phase_proc_lp seems to have issues with IRIS RAW files. I am trying to use this function to recalculate KDP values, but as it can not correctly unfold the PhiDP values, it affects the recalculation of KDP as well. There are no issues in rays with no folding of PhiDP, but in rays where folding occurs it is problematic: it starts with a negative value and the highest value in these rays is 0 degrees.
The original PhiDP data with folding in some rays [image: phidp_orig_03072016_zoom] https://user-images.githubusercontent.com/12828811/32225493-1939182c-be4e-11e7-96b7-8deb52ab596a.PNG
Unfolded PhiDP using phase_proc_lp [image: phidp_corrected_03072016_zoom] https://user-images.githubusercontent.com/12828811/32225529-2e32f78e-be4e-11e7-8c9b-5405a56561a9.PNG Recalculated KDP has similar signature as a result: [image: kdp_corrected_03072016_zoom] https://user-images.githubusercontent.com/12828811/32225643-a79c548a-be4e-11e7-9a8b-0ca8ac1e7938.PNG
Playing around with nowrap and overide_sys_phase parameters did not seem to help either. The first did not have any effect seemingly and the second one changed all the values, but the unwanted behaviour resisted (as was expected).
As a quick and dirty fix I can think of adding a check for each ray which reads the minimum negative PhiDP value (if present) and adds this to each bin in this ray.
To check the original file I uploaded it here (NB! the file has original PhiDP saved in "reverse", to fix it one should change the sign and add 180 to PhiDP original data, e.g. [orig_phidp*(-1)+180] and the KDP is negative) SUR160703101507.zip https://github.com/ARM-DOE/pyart/files/1430819/SUR160703101507.zip
Has anyone noticed something similar with their files? Do you have any suggestions why this is happening?
— You are receiving this because you are subscribed to this thread. Reply to this email directly, view it on GitHub https://github.com/ARM-DOE/pyart/issues/695, or mute the thread https://github.com/notifications/unsubscribe-auth/AAyYB92NvMA6Jll0-ee9BJ5EZ4Ih0DuBks5sxyJtgaJpZM4QMv85 .
I know it's an old issue but, I've done what you suggested @scollis : shoe-horning the differential phase data into the region based dealiaser, and it works very well!!! Here step by step:
- Raw differential phase:
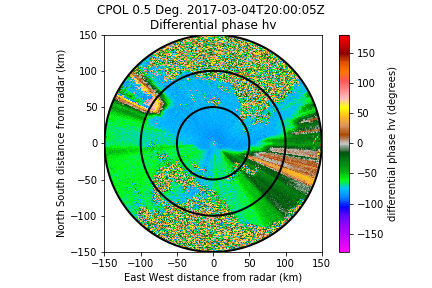
- Use of region based dealiaser (I put the argument nyquist_vel to 90 degrees), and applied a 90 degrees offset to put the data to zero:

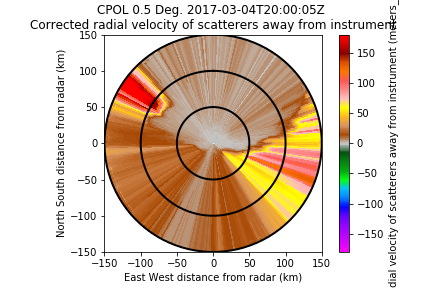
- Use of phase_proc_lp to dealias step 2:
And for comparison what you get if you directly try to unfold the raw differential phase:
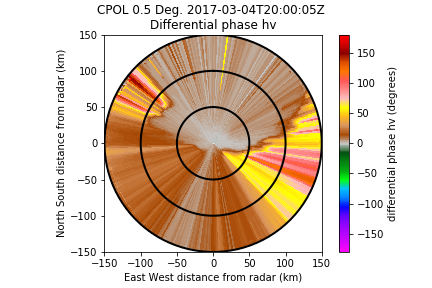
I've tried it on several files, it is much better to use the region based dealiasing first.
Fantastic! Perhaps we can work up a pr when we visit. Very encouraging
-sent from a mobile device-
On Oct 14, 2018, at 9:24 PM, Valentin Louf [email protected] wrote:
I know it's an old issue but, I've done what you suggested @scollis : shoe-horning the differential phase data into the region based dealiaser, and it works very well!!! Here step by step:
Raw differential phase:
Use of region based dealiaser (I put the argument nyquist_vel to 90 degrees).
Use of phase_proc_lp to dealias step 2. (and applied a 90 degrees offset to put the data to zero):
And for comparison what you get if you directly try to unfold the raw differential phase:
I've tried it on several files, it is much better to use the region based dealiasing first.
— You are receiving this because you were mentioned. Reply to this email directly, view it on GitHub, or mute the thread.
@scollis I can't remember, but was this ever added?