Registration of two FA images - Output is flipped
Hello,
I am trying to nonlinearly register two FA images (derived from diffusion MRI). I am particularly interested in the affine map and warp nifti file that ANTspy registration generates. I apply the transformations and save my moved file. when I load the file in any other viewer (eg: MI-Brain), the output FA image is flipped and not correctly aligned with the fixed FA file.
Here's how I load data and call the registration function:
import nibabel as nib
import ants
import numpy as np
ni_img1 = nib.load(path)
fixed = ants.nifti_to_ants(ni_img1)
# fixed = fixed.reorient_image2() #RAS
ni_img2 = nib.load(n_path)
moving = ants.nifti_to_ants(ni_img2)
# moving = moving.reorient_image2() #RAS
mytx = ants.registration(fixed=fixed , moving=moving, type_of_transform='SyN' )
mywarpedimage = ants.apply_transforms(fixed=fixed, moving=moving,
transformlist=mytx['fwdtransforms'])
I think the problem could be that my data is in RAS and ants converts it into LAI space. Here are the specs for the images.
fixed ANTsImage (LAI) Pixel Type : float (float32) Components : 1 Dimensions : (182, 218, 182) Spacing : (1.0, 1.0, 1.0) Origin : (90.0, -126.0, -72.0) Direction : [-1. 0. 0. 0. 1. 0. 0. 0. 1.]
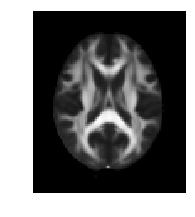
moving
ANTsImage (LAI)
Pixel Type : float (float32)
Components : 1
Dimensions : (116, 116, 72)
Spacing : (1.9828, 1.9828, 2.0)
Origin : (115.0, -100.8727, -80.4458)
Direction : [-1. 0. 0. 0. 1. 0. 0. 0. 1.]
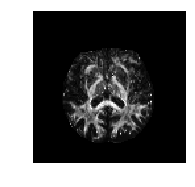
mywarpedimage ANTsImage (LAI) Pixel Type : float (float32) Components : 1 Dimensions : (182, 218, 182) Spacing : (1.0, 1.0, 1.0) Origin : (90.0, -126.0, -72.0) Direction : [-1. 0. 0. 0. 1. 0. 0. 0. 1.]

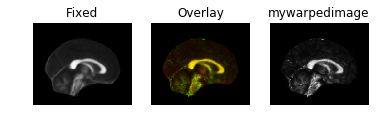
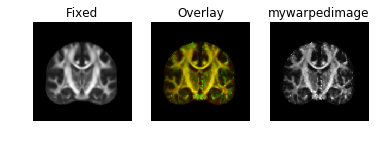
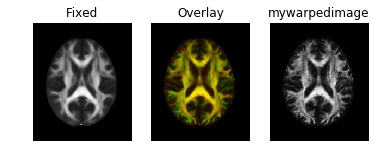
Results look good when visualized inside the same notebook. However, when I save the warped image to nifti file, it does not align in any other viewer. (I tried saving as ants image and also like this mywarpedimage.to_nibabel() but in all cases same results)
Here's how it looks: Fixed image:
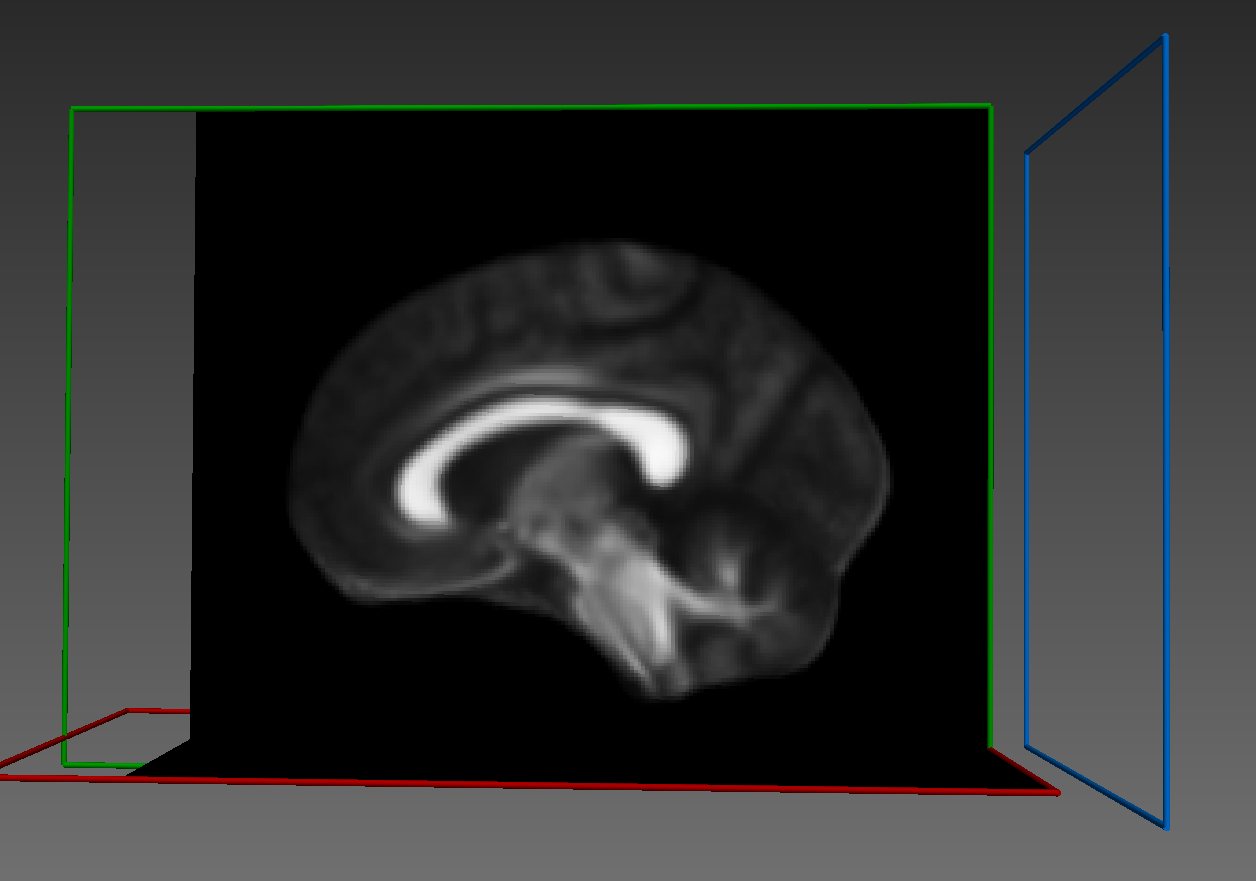
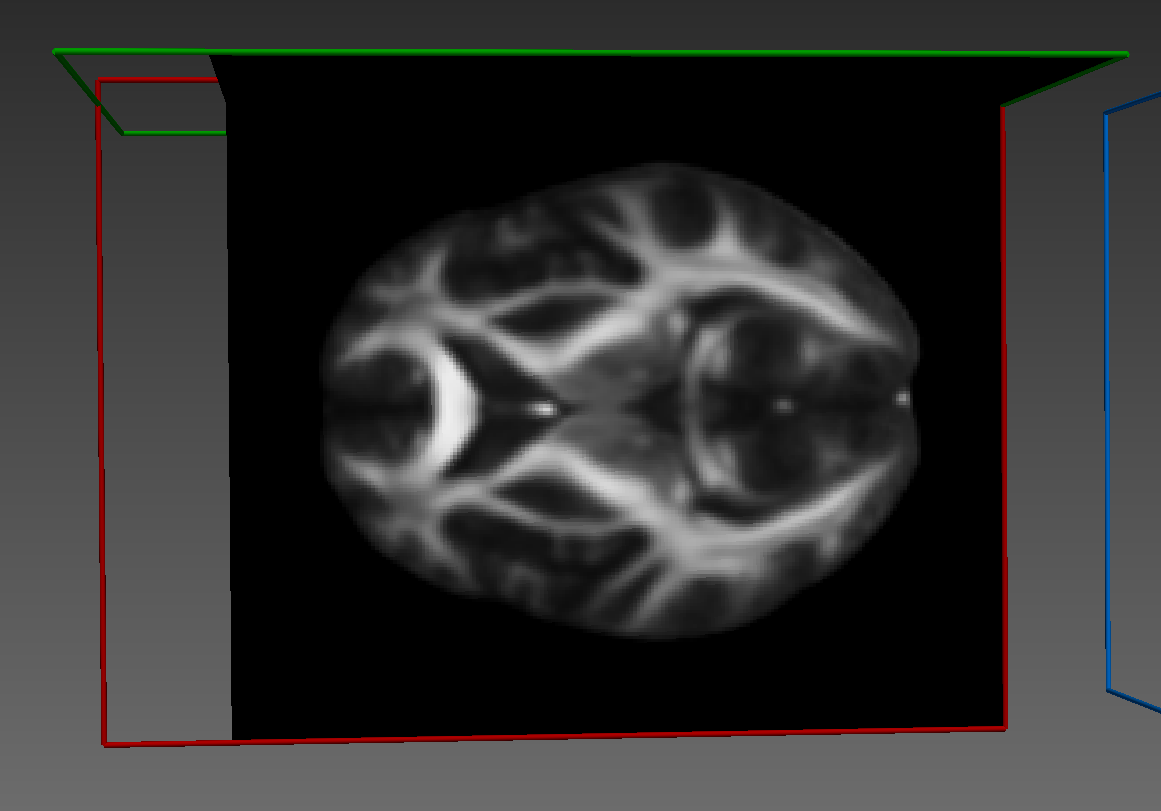
Warped image:
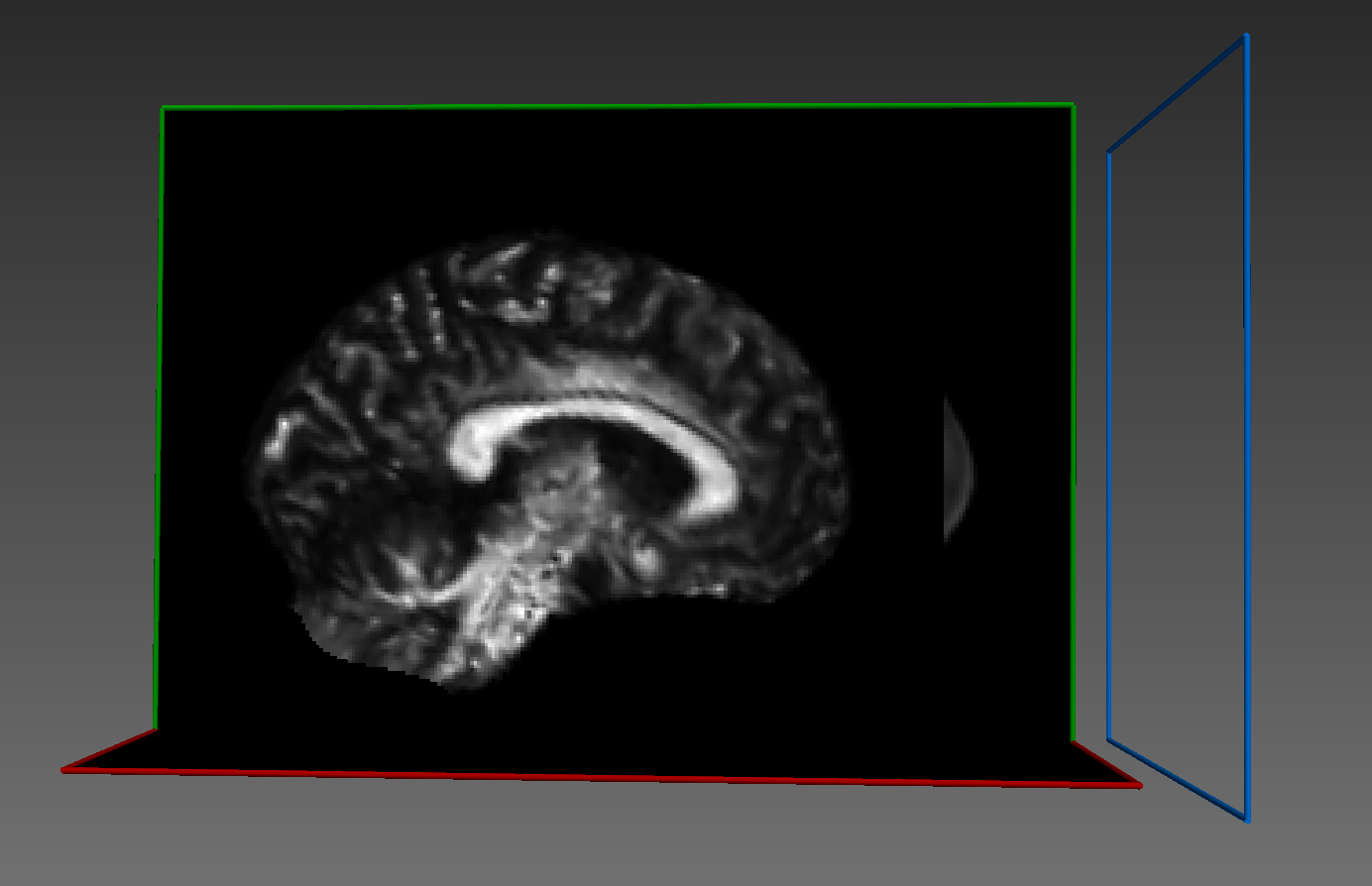
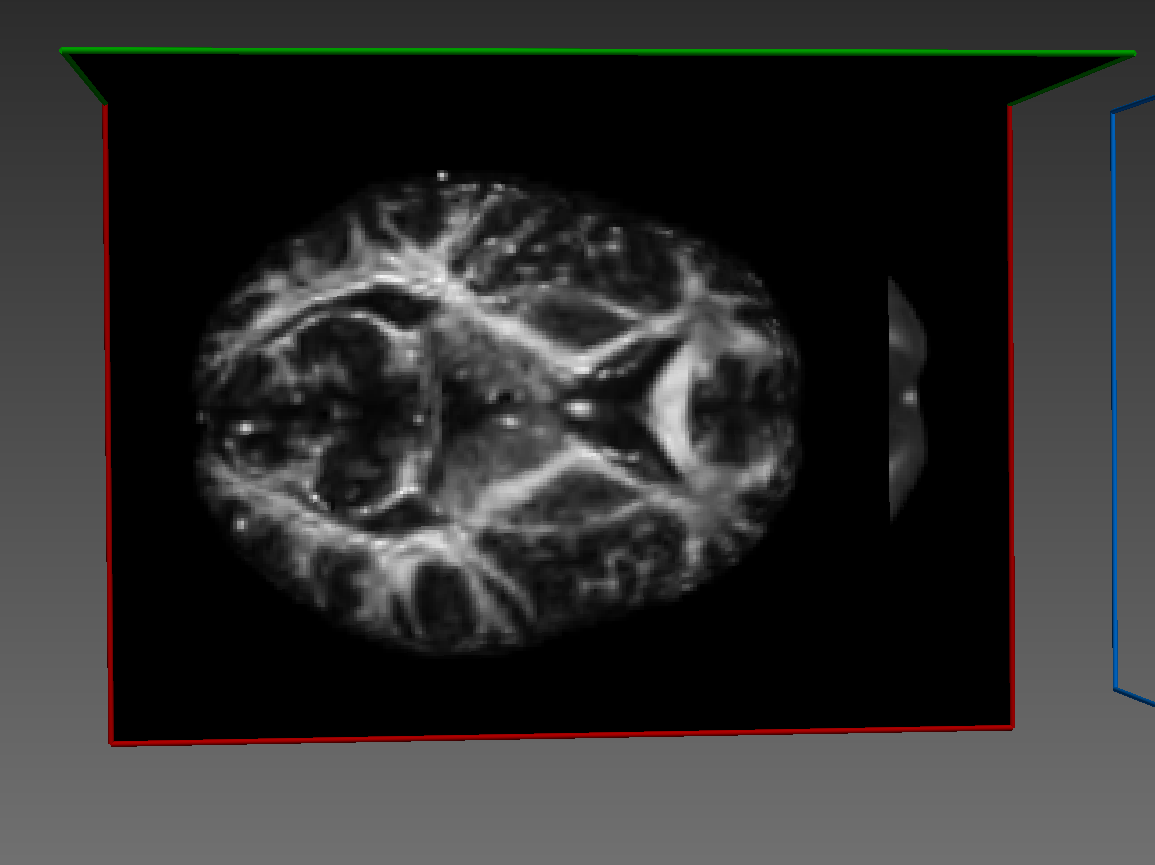
Some sort of flipping. I tried changing the co-ordinate system of ants images as you can see commented lines in the code above. I tried using reorient_image2() function to have data in RAS before registration or even after registration. But in all cases, it gives me the same results.
On a side note, I tried everything, registering two images of the same size, dimensions, voxel sizes, etc. In all cases, my final output is flipped or not in the same space.
I am most interested in the transformation maps. I need them to be in the right space. Do you know what would be the solution? am I doing something wrong here?
Thank you very much for the help!
Different packages use different elements of the nifti header in different ways. It's unfortunate but we don't have the capacity to facilitate compatibilities with these other packages. Know that transforms and orientations are mutually consistent within the ITK ecosystem (i.e., ANTs, ITK-SNAP) and so, from that perspective, they are in the right space.
Hi @ntustison
I found the solution.
Instead of loading data with nibabel and then converting it into ants image like this:
ni_img1 = nib.load(path)
fixed = ants.nifti_to_ants(ni_img1)
fixed = fixed.reorient_image2() #RAS
I load data directly with ants API using ants.image_read like this:
fixed = ants.image_read(path)
fixed = fixed.reorient_image2() #RAS
It worked like a charm ;)
I think when we load data with nibabel and convert it into ants image, we might be losing some information about the data.
The nifti_to_ants function gets the qform from the source image and then uses it to set the antsImage direction matrix.
image_read, as far as I can see, uses ITK I/O, which uses the sform if available. If your image contains both and they aren't the same transform, it's possible to get different orientations.
To make things more complicated, ITK has changed over time, it used to prefer qform (which may be why nifti_to_ants does likewise) but now prefers sform.