LesionFilling error
Dear ANTs experts:
I have been trying to fill a T1-weighted image with a white matter lesion mask and I encountered the error shown below:
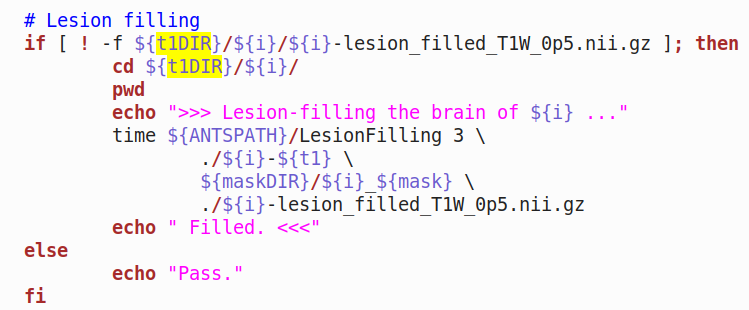
The relevant portion of the BASH script used:
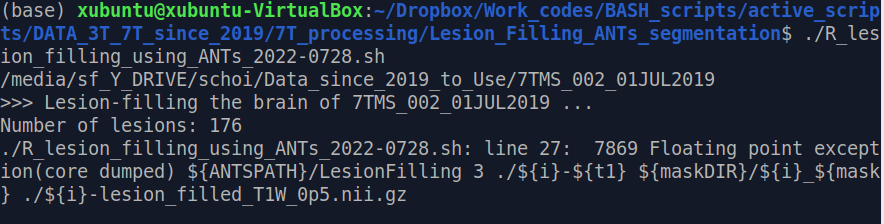
Attempt using R I tried it again with ANTsR and I encountered an error message shown below:
R code: t1DIR = "/media/sf_Y_DRIVE/schoi/Data_since_2019_to_Use" maskDIR = "/media/sf_Y_DRIVE/DATA_from_collaborators/from_Rong/ms7t_data_share/agg_report" scancode = "7TMS_002_01JUL2019" img1 = paste0(t1DIR,"/",scancode,"/",scancode,"-pre-denoised-T1W_0p5.nii.gz") img2 = paste0(maskDIR,"/",scancode,"_wml_gt.nii.gz") t1w = antsImageRead(img1, 3) wml = antsImageRead(img2, 3) filled <- basicInPaint(t1w, wml)
ITK ExceptionObject caught ! Error in fastMarchingExtension(speedimage, healthymask, img) : /home/xubuntu/R/x86_64-pc-linux-gnu-library/3.6/ITKR/libs/include/ITK-5.2/itkFastMarchingImageFilterBase.hxx:320: itk::ERROR: itk::ERROR: FastMarchingExtensionImageFilterBase(0x563566d54480): Discriminant of quadratic equation is negative
Any suggestions or advice will be appreciated! Best, SC