relion_display fails on multiview with lowpass
When I display multiple micrographs from a .star file with relion_display, adding --lowpass causes a constant value image to be displayed instead of the filtered image. This appears to have been happening for at least three years per a CCPEM thread. Correcting this will presumably be prerequisite to addressing #373.
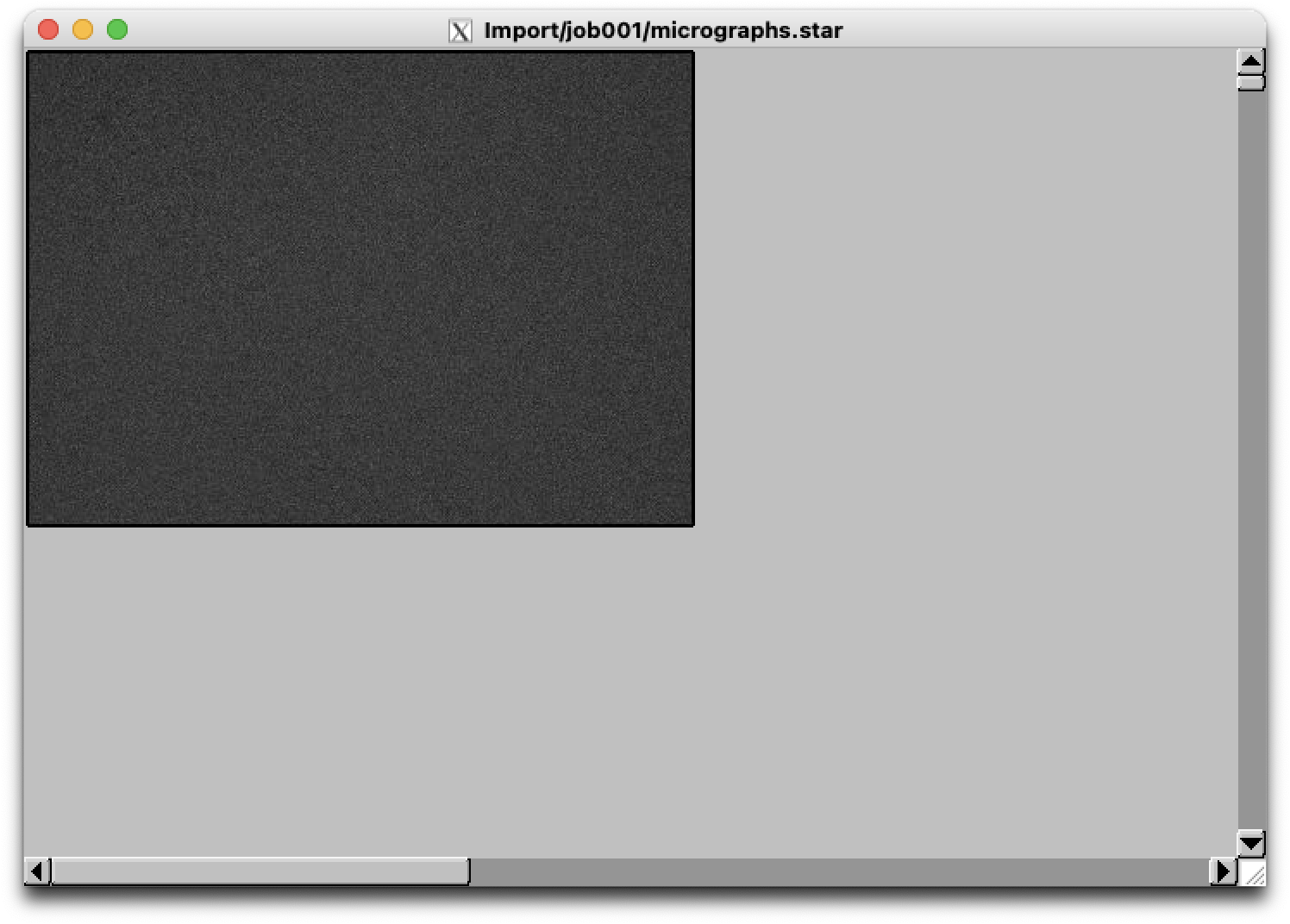
For example, running without lowpass yields an image:
$ relion_display --scale 0.067 --max_nr_images 1 --i /cephfs/alewis/relion_projects/rho2/Import/job001/micrographs.star --display rlnMicrographName --angpix 1.3398
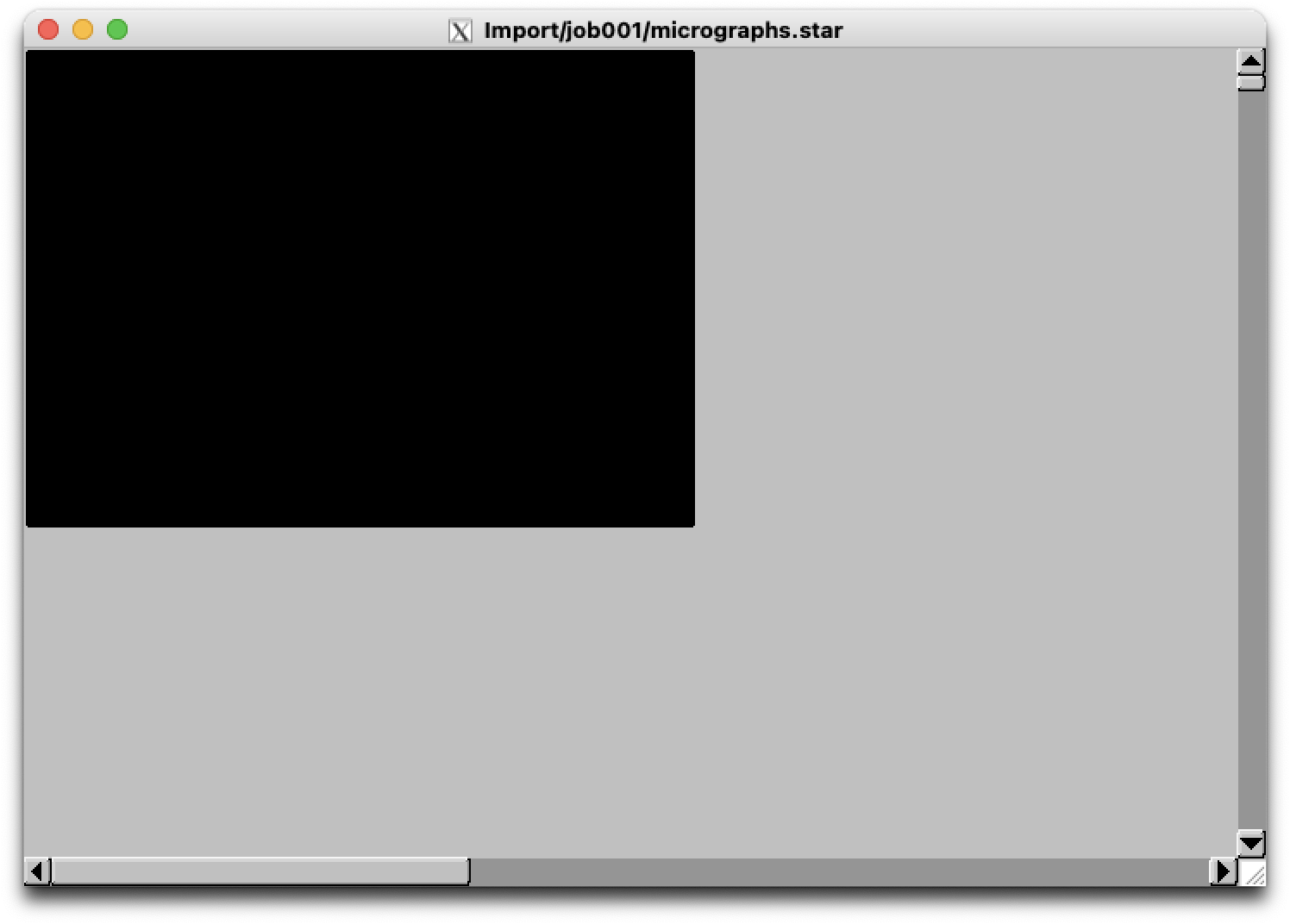
whereas running with lowpass does not:
$ relion_display --scale 0.067 --max_nr_images 1 --i /cephfs/alewis/relion_projects/rho2/Import/job001/micrographs.star --display rlnMicrographName --angpix 1.3398 --lowpass 20
I've included absolute paths above so that anyone at the LMB wanting to reproduce the issue on the same micrograph can do so, at least in the short term.
Environment: LMB hex sourcing relion-4.0-dev-slurmified.csh.
- OS: Scientific Linux 7.9
- MPI runtime: OpenMPI 2.0.1
- RELION version: 4.0-beta-2-commit-726d82
- Memory: 3.17 TB
- GPU: 2x Tesla T4
Dataset:
- Micrographs: K3 (3456 x 4092 pixels)
- Pixel size: 1.3398
Interesting. Apparently this was repaired in #247 but then broken again at some point. I reproduced the issue.
I myself won't work on this, however, because I am against opening hundreds or even thousands of micrographs on relion_display. It takes too long and consumes too much memory. I recommend using the Manual Picker GUI as mentioned in the CCPEM message.
I leave this issue open so that others can work on this.
Unfortunately the manual picking GUI is not useful for deciding which micrographs to discard, because it does not provide thumbnails. Clicking "Pick" to open each micrograph, wait for it to load, close it and then toggle the corresponging radio button would end up being vastly slower than using relion_display (which one can leave to load in the background while doing something else). I'd be curious if there's a viable alternative (besides simply doing it in cryosparc).
I don't visually select good and bad micrographs. I just process everything and use Class2D and/or 3D to discard bad particles. I have never seen convincing cases where manual curation of micrographs helped. (Do you really do this for > 10,000 micrographs?)
I do. It usually takes <5 minutes to page through 10k mics, tossing maybe 300 due to ice thickness/fractures/contamination/missing graphene support/etc. It seems reasonable to take easy steps to clean up the data, since classification is an imperfect filter. Another reason to use cryosparc I suppose.
Does it improve the resolution?
I haven't tried to quantify that, or seen any reports; what I do know is that many methods sections describe manually inspecting mics for defects, as does issue #373 and the CCPEM thread. I'd volunteer to try tossing bad mics from a finished processing project and see if it does help, if relion_display were working. But regardless the answer will be dataset-dependent.
I know some people do it, but as I wrote above, "I have never seen convincing cases where manual curation of micrographs helped" (based on my own experiences of reprocessing many, diverse datasets from EMPIAR and my colleagues' experiences). Unless proven, I consider this practice worthless. In old days, some people visually curated individual particles but most of them no longer do so because it turned out unnecessary. I think micrograph curation is the same.
That would certainly be convenient. One specific trade-off I don't know how to weigh arises when ~1% of the micrographs have a small (~1-5) number of particles (e.g. holes missing graphene oxide or carbon support). In that case RELION warns the SNR model will be unstable and recommends regrouping. I would hesitate to regroup all the micrographs just to correct an issue arising from micrographs that could be discarded outright. But I don't know if there is a downside to regrouping, or how much the SNR model instability would affect a final model if left unaddressed entirely. The mismodeled particles would be rare (~0.01% of the total), but might an overestimated SNR cause them to contribute a disproportionately large amount of error to the reconstruction? Simply discarding them always seemed simplest, but maybe you'd know if either or both alternatives is just as good.
As long as the refinement runs fine, you can safely ignore the warnings.
might an overestimated SNR cause them to contribute a disproportionately large amount of error to the reconstruction
I cannot say this never happens but in practice this is very very unlikely.
You can also remove suspicious micrographs with few particles (as you mentioned) or those with very low scale factors (see *_model.star).
Sounds good, thanks!