sgkit
 sgkit copied to clipboard
sgkit copied to clipboard
Some results of Allele Frequency Distributed Benchmark
Here is some results of Allele Frequency's distributed benchmark on 1000 Genome data:
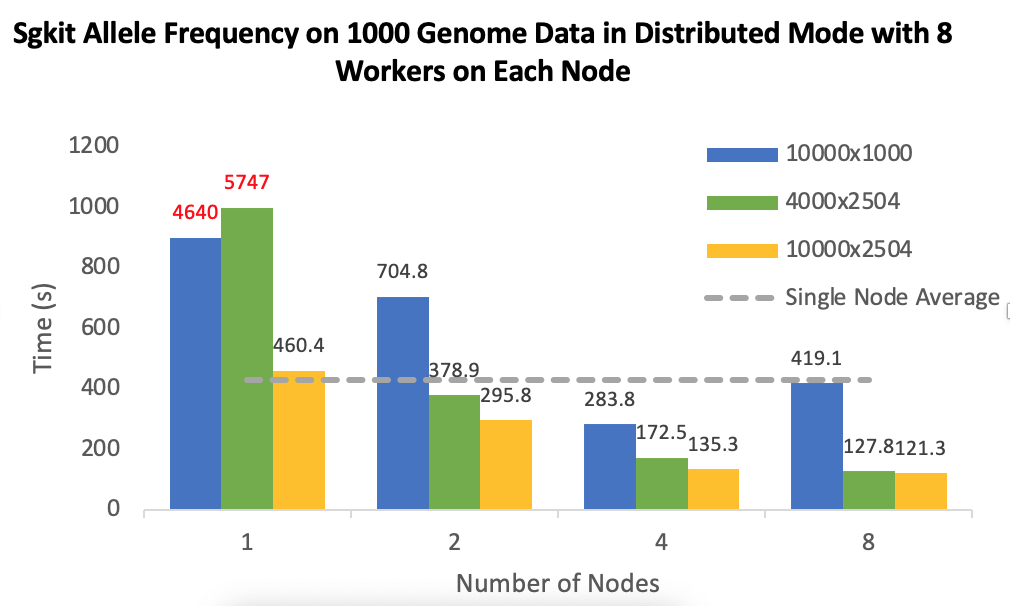
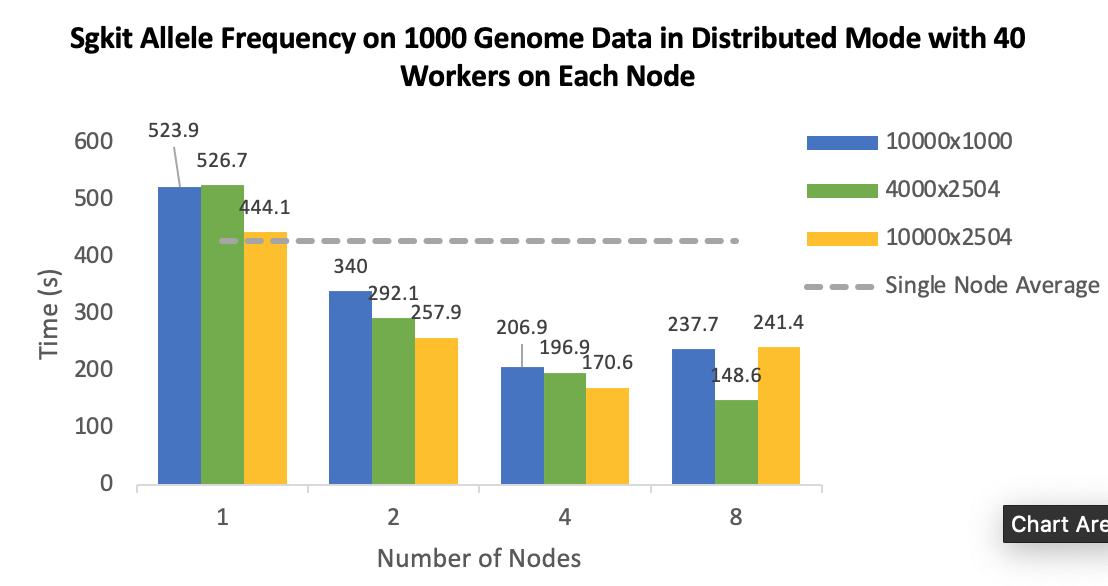
I set up the experiment with [1,2,4,8] machines, data are stored in HDFS running on the nodes respectively, block size is 64 MB.
Data chunk settings: The default chunk size of data is 10000x1000(Variants x Samples), so basically sharding in tiles. I also do experiments on row sharding: 4000x2504, as it has the same size of each chunk with default sharding, 10000x2504 to explore how sizes of chunks affect runtime. I tried 1000x2504 as well, but it seems that workers die silently in this case. I believe Hail also do row shardings, so genotype data of a specific variant from all samples appear in one chunk/partition.
Dask scheduler: Only the gray lines in the plots, "Single Node Average", uses single machine scheduler, this is data from single node experiment to be used as a baseline in distributed experiments. I think the single machine scheduler was created in the same process as Sgkit program running, also the worker is created in that same process as well. I believe it has 1 worker 40 threads. The other experiments and runtime are from distributed scheduler.
Dask workers settings: Because each machine I'm using have 40 vCores, I primarily tested two settings:
- 8 workers on each machine, each worker has 5 threads. This is the auto mode configuration (dask-worker --nprocs auto). Note this is not the default mode, the default is 1 worker 40 threads, but will run endlessly on 1 or 2 nodes.
- 40 workers on each machine, each worker has 1 threads. Because this setting seems perform well in 1/2 nodes
The following plot is performance with maximum computation resource I get (8 nodes, 320 vCores, 160 physical cores):
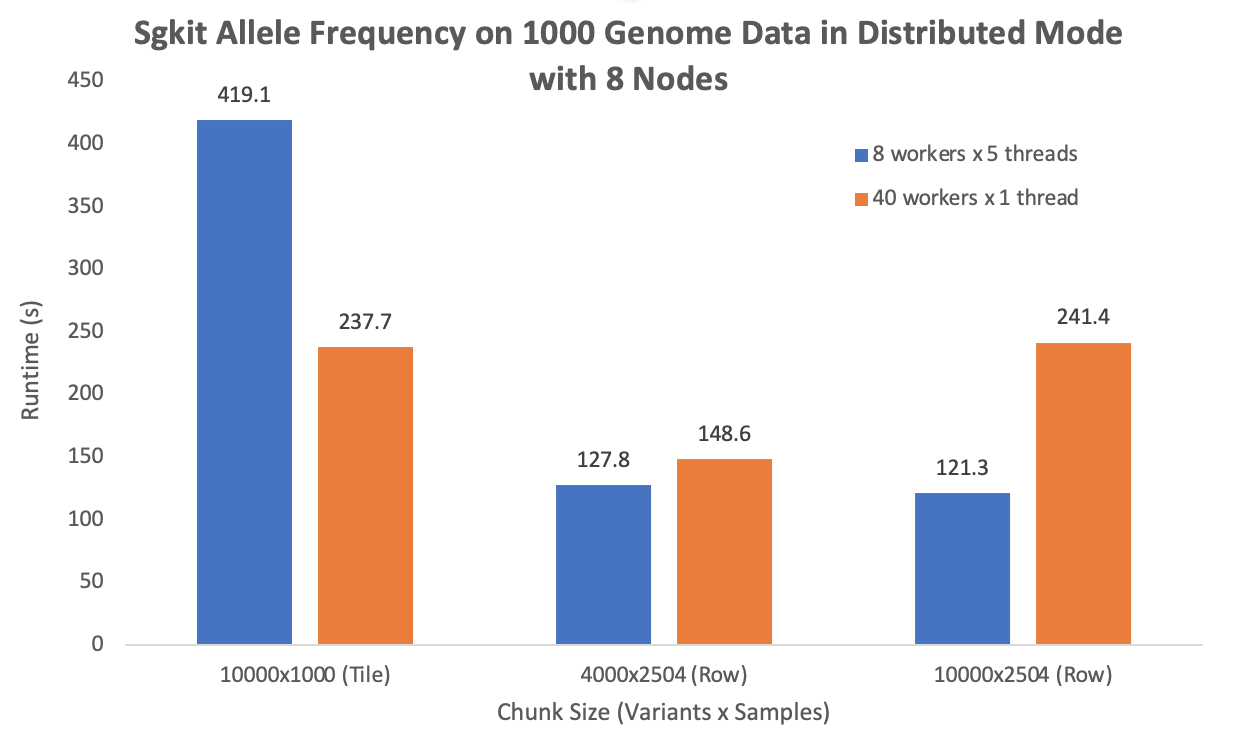
Very nice @LiangdeLI, this is an informative analysis!