rootstock
 rootstock copied to clipboard
rootstock copied to clipboard
Can't browse pandoc-crossref subfigures using Lightbox
This comment references #191 where there are more details.
Viewing the figures on their (Lightbox?) only shows plain figures, not the subfigures with pandoc-crossref. In the screenshot, only the four non-subfigures are shown (note the figure numbering indicates other figures in the document that can't be browsed).
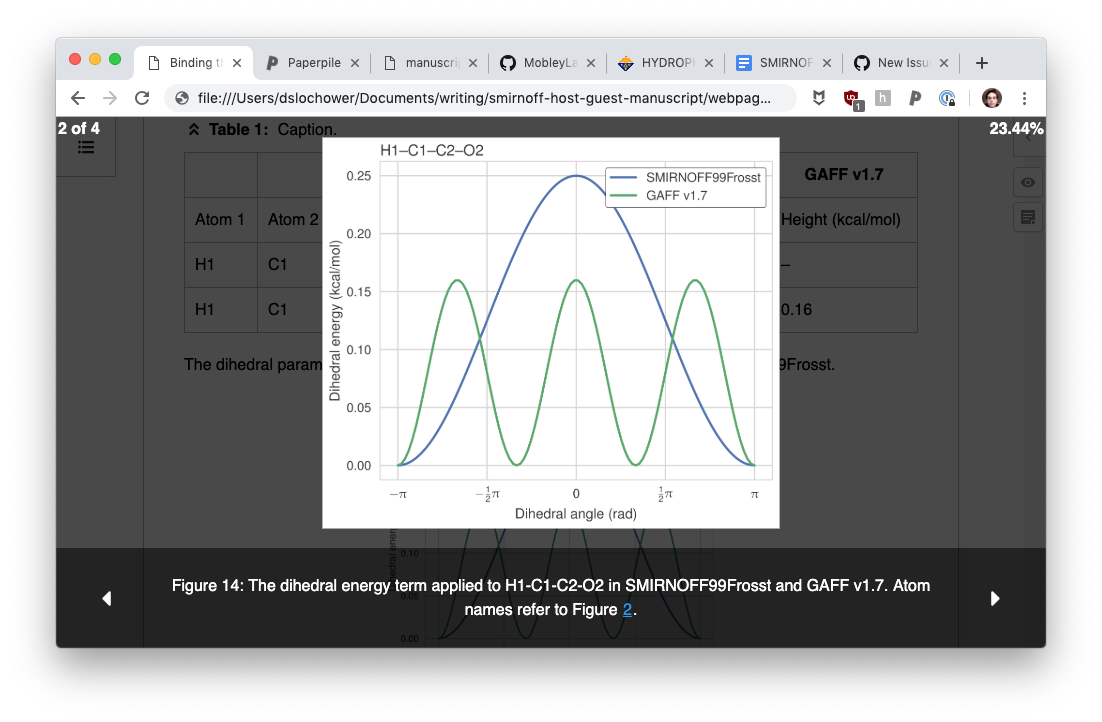
I am guessing this is happening because the subfigures that use pandoc-crossref, do not use the HTML <figure> syntax. Inferring from this HTML, pandoc-crossref inserts:
<div id="fig:atom-names" class="subfigures" data-collapsed="false">
<p>
<img src="images/atom-names-trimer.png" title="fig:" alt="a" style="width:3.5in">
<img src="images/gaff-atom-types.png" title="fig:" alt="b" style="width:3.5in"></p>
<p>Figure 2: Atom names (left) and GAFF atom types (right) for a glucose monomer in αCD shown with two flanking monomers. The remaining three glucose monomers are hidden for clarity.. </p>
</div>
Perhaps @vincerubinetti would be able to detect that this is a figure using the class="subfigures"?
@dhimmel That's right. Though detecting the subfigures seems like a brittle solution. We'll have to sit down and think about how to solve this in the best way. We'll have to experiment with multiple different plugins and html structures to see if we can stress test it.
Would it work to add subfigures here, as well?
https://github.com/manubot/rootstock/blob/091ca8d85c8ef2d7af16fcc8d2ed3ebcbc187f13/build/plugins/lightbox.html#L46
Would it be easy to add a button to collapse long tables? I'm primarily asking here to coalesce the aesthetics questions for @vincerubinetti .
I think it would also be super spiffy to have an ability to directly export tables as CSV (kind of a wishlist feature, probably). But I hunt through SI so many times looking for machine readable tables.
@vincerubinetti Have you thought any more about a non-brittle solution for subfigures here and in #207? If not, if you can point me in the right direction, I'd like to make HTML class="subfigures" behave the same way as normal figures and I'm happy to employ some prototype solutions in my working manuscript. I think having subfigures autohyperlinked but not browsable by tooltip or with Lightroom has confused a few readers.
For example: hovering over "Figure 3" shows Figure 11 in my manuscript, clicking on Figure 3 shows the top of the manuscript, and none are shown in Lightbox.

https://slochower.github.io/smirnoff-host-guest-manuscript/
I can give some suggestions for fixing this in the short term, but the longer-term/better solution will take some thinking.
tooltips problem
The problem is with these lines: https://github.com/manubot/rootstock/blob/master/build/plugins/tooltips.html#L401-L415
This is the function that goes to whatever the citation/link points to (be it a figure, reference, etc) and gets that thing so it can be shown inside the tooltip. For figures, it gets the parent node of the thing, because normally the html is
<figure>
<img id="fig:some-figure"/>
<caption>caption text</caption>
</figure>
and we want to include the caption text too.
In your case, the html looks like
<body>
<div id="fig:some-figure">
... other stuff ...
</div>
</body>
so the plugin ends up copying the entire body of the document into the tooltip.
Change the function to this:
// get element that is target of link or url hash
function getSource(link) {
const hash = link ? link.hash : window.location.hash;
const id = hash.slice(1);
let target = document.querySelector(
'[id="' + id + '"], [name="' + id + '"]'
);
if (!target)
return;
let newTarget = target;
// if figure or table, modify target to get expected element
if (hash.indexOf('#ref-') === 0)
newTarget = target.querySelector('p');
else if (hash.indexOf('#fig:') === 0)
newTarget = target.parentNode;
else if (hash.indexOf('#tbl:') === 0)
return;
if (!newTarget || newTarget === document.body)
newTarget = target;
return newTarget;
}
Note that this is slightly sloppy, but it should work for now. All this does is add a check that the target is not the body of the document, and that it's not some other error or undefined value.
Great @vincerubinetti! That's working for the tooltips! Thanks so much. (Haven't pushed the changes to GitHub yet, so it's not live on the public-facing version yet).
I think something very similar is going on with the hot links. The figure cross-references link to anchors on the page, like so: https://slochower.github.io/smirnoff-host-guest-manuscript/#fig:dG-dH (This is correct.) Here's the unexpected thing: if you open this URL directly in a new window, the page will load at the appropriate figure. But if you click the hyperlink from the main document, the link sends you to the top of the page.

This may also be related to the Lightbox issues :)
lightbox problem
Change
const imgs = document.querySelectorAll('figure > img');
to
const imgs = document.querySelectorAll('img');
to get all images in the document instead of just those that are direct children of <figures>.
Change
bottomContainer.appendChild(caption);
to
if (caption)
bottomContainer.appendChild(caption);
to make sure function doesn't crash if caption text can't be found.
Change
const captionSource = img.nextElementSibling;
to
const captionSource = img.nextElementSibling;
if (!captionSource || captionSource.innerText.trim() === '')
return;
to return undefined (to signify caption wasn't found) if the element after the image doesn't exist or doesn't contain any text.
Again, this is a quick and dirty solution just to help you out for the time being. The real solution will be more generic, and probably more complicated.
The Lightbox improvements help a lot! You're right that a generic solution may be more complicated.
- These changes pick up stuff like the Twitter and ORCID icons next to author names.
- These changes split up the subfigures, so that each panel is shown separately (Lightbox says XX of 83) in the top left, instead of the ~27 total figures. I will also independently look into ways of combining the subfigures into a single figure.
Thanks again. I appreciate it. Finally, is it the jump-to-first.html plugin that is changing the behavior of the anchored links?
Thanks again. I appreciate it. Finally, is it the
jump-to-first.htmlplugin that is changing the behavior of the anchored links?
Can you elaborate on this? Not sure what behavior you're talking about.
Oh, sure.
I tried to explain in https://github.com/manubot/rootstock/issues/209#issuecomment-489717072. But maybe it's easiest to demonstrate. If you go here: https://slochower.github.io/smirnoff-host-guest-manuscript/ and click on the first text instance of "Figure 3" (second paragraph, results section) it jumps to the top of the manuscript. (This happens even on my local copy, where I've updated tooltips.html and lightbox.html per your guidance.) The link is formatted correctly -- it has a # anchoring element at the end -- and it works if you go directly to the link in a new tab, but not if you click from within the main manuscript page.
Oh okay, I didn't realize you were referring to that specifically.
Honestly this subfigure thing breaks so many features. That particular issue seems somewhat minor. Perhaps we can address the more pressing issues, like the tooltips and lightbox ones, just so you can have it fixed for your manuscript? Fixing all of these proper will take some time. The issue is at least documented here now, and I can come back to it when I have time.
Sure, that's totally understandable.
I think the root of the issues is the use of pandoc-crossref. I just added a comment on pandoc-fignos to see if there's been any development on subfigure support (https://github.com/tomduck/pandoc-fignos/issues/50). I'm guessing not.
Because the broken links have been bothering some of my coauthors, there are two immediate solutions that would enable me to revert to pandoc-fignos:
- Have the analysis pipeline compose the subfigures directly (this is not very feasible in my case because the source directory has hundreds of different figures).
- Use a command line tool like ImageMagick's montage to place the figures on a grid and save as a single file.
These options add an intermediate step between the data analysis and the manuscript, which is not ideal, but maybe we could write a pandoc filter to take care of this automatically in the future.
Use a command line tool like ImageMagick's montage to place the figures on a grid and save as a single file.
This seems like a good solution to me. On one hand, I think subfigures make a lot of sense theoretically. For example, it's helpful sometimes to be able to copy subfigures individually. On the other hand, I am not aware of any journals whose interface supports subfigures. Thus, I am guessing at some point, to deal with a journal, you will have to switch to combined figures. I am also not sure of subfigures exportability to various formats like JATS or DOCX. So I think they may end up being a can of worms that is best to just avoid for the time being.
On the other hand, I've endured much agony trying to make paneled figures, especially when combining SVGs. So being able to defer the work to subfigures may be nice.
This seems like a good solution to me.
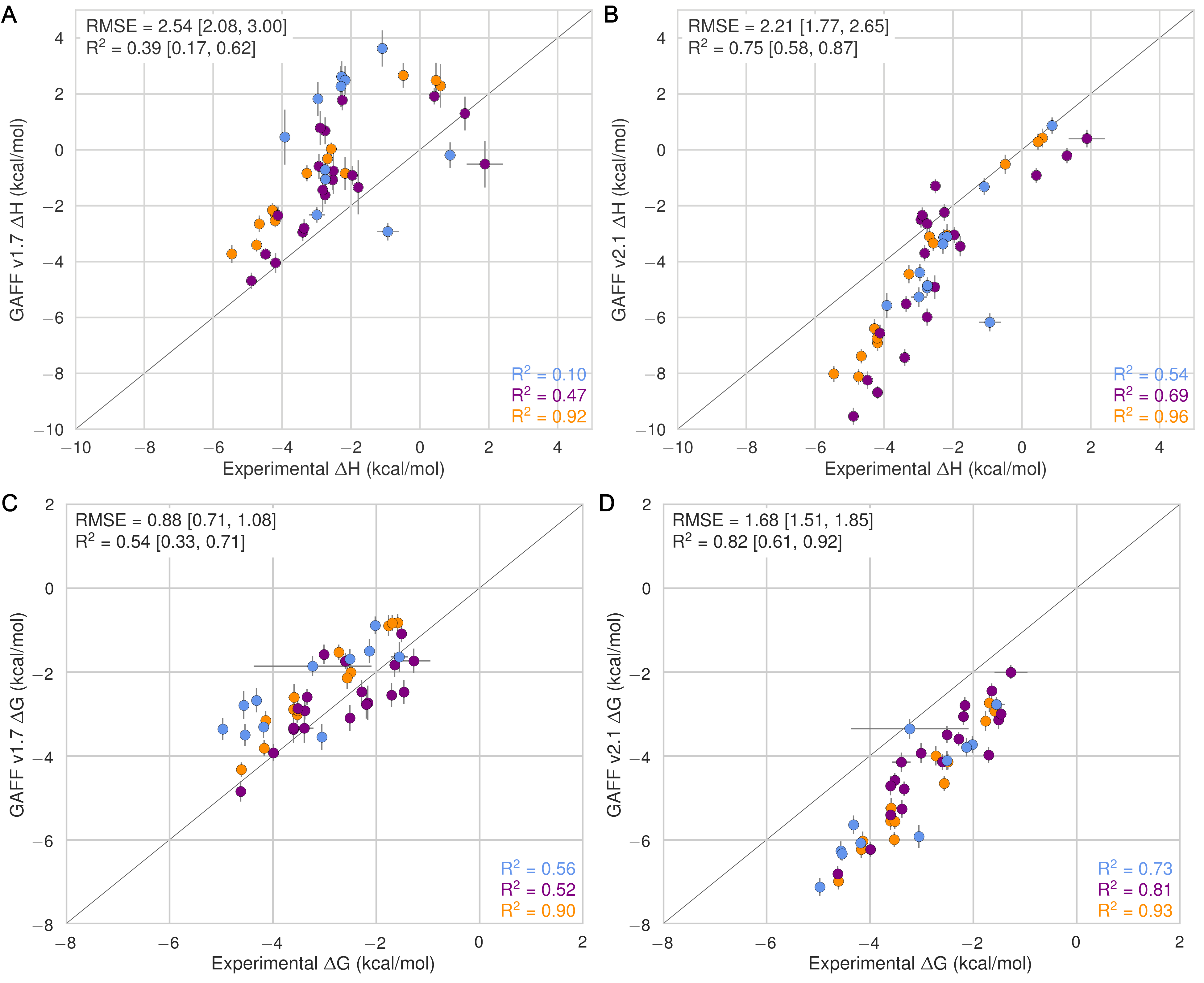
Dropping this pseudocode here, in case it is useful for future reference. This is obviously not robust yet, but contains the basic ImageMagick commands we might want to use.
import os
import subprocess
figures = ["../images/GAFF-v1.7-vs-Experiment-dH.png",
"../images/GAFF-v2.1-vs-Experiment-dH.png",
"../images/GAFF-v1.7-vs-Experiment-dG.png",
"../images/GAFF-v2.1-vs-Experiment-dG.png"]
def int_to_letter(int):
alphabet = list('abcdefghijklmnopqrstuvwxyz'.upper())
return alphabet[int]
def label(input, output, annotation):
command = f"""
convert \
-font arial \
-stroke black \
-pointsize 18 \
-gravity northwest \
-draw "text 2,2 '{int_to_letter(annotation)}'" \
-density 300 \
{input} \
"tmp/{output}"
"""
subprocess.call(command, shell=True)
def tile(output, geometry="+2+2"):
command = f"""
montage \
tmp/tmp-?.png \
-geometry {geometry} \
tmp/{output}
"""
subprocess.call(command, shell=True)
def clean(mask):
command = f"""
rm tmp/{mask}*.png
"""
subprocess.call(command, shell=True)
for index, figure in enumerate(figures):
if not os.path.exists("tmp"):
os.makedirs("tmp")
label(input = figure,
output = f"tmp-{index}.png",
annotation = index)
tile(output = f"rendering.png")
clean(mask = "tmp")
Attached a rendered example:
Agree with the rest of what you've said, @dhimmel .
Edit: have not tested on SVG or PDF, but expect those won't work with this code.