dffml
 dffml copied to clipboard
dffml copied to clipboard
gsoc: docs: intuitive: Deploy JupyterLite
Understand where interfaces are for opening and saving files
Check Jupyter adaptability How does Jupyterlite get deployed as static site Can I run it from Github pages? Figure out where open file and save file code is
JupyterLite runs with Pyolite - https://jupyterlite.readthedocs.io/en/latest/kernels/pyolite.html
Pyolite is built on top of Pyodide (CPython to WebAssembly) - https://github.com/pyodide/pyodide
Pyodide makes it possible to install and run Python in the web browser using micropip - https://pyodide.org/en/stable/usage/api/micropip-api.html
https://github.com/jupyterlite/jupyterlite/blob/main/app/lab/index.template.js https://gist.github.com/jtpio/3763af75ec8cd7bfdce86501563fef09

Link: https://github.com/jupyterlite/jupyterlite

Link: https://jupyterlite.readthedocs.io/en/latest/architecture.html
- https://jupyterlite.readthedocs.io/en/latest/quickstart/deploy.html
- How to start working with jupyterlite
- https://intel.github.io/dffml/master/examples/notebooks/moving_between_models.html
- This is one of the Notebooks that have already been written for DFFML. Parent page with all Notebooks: https://intel.github.io/dffml/master/examples/notebooks/index.html
- https://clairefollett.github.io/jupyterlite_demo/lab/index.html
- Claire's github pages version of jupyterlite demo
- [x] Updated pip, setuptools, wheel
- https://github.com/intel/dffml/blob/main/docs/contributing/dev_env.rst#virtual-environment
- [x] Created a new directory to store code for GSoC 2022
~/gsoc2022- https://developers.google.com/open-source/gsoc/timeline
- [x] Recording with
asciinema - [x] Created virtual environment
python -m venv .venv
- [x] Activated virtual environment
Instead of targeting rst files, we're going to work with already existing Notebooks
- Devtools
- Found the Upload File button in jupyterlite
Next steps:
- Grab the jupyterlite source code
- https://jupyterlite.readthedocs.io/en/latest/contributing.html#get-the-code
- Look for where in the javascript they have the file upload
git grep "Upload File"
- Figure out, how would you use the console with devtools, to trigger the file upload mechanism
- Investigate how to trigger the file uploaded mechanism (looks like a regular
<input>element) - https://stackoverflow.com/a/66466855/12310488
- Do not use the answer that has the green check, it is not relevant to our use case, see the one linked.
- Investigate how to trigger the file uploaded mechanism (looks like a regular
Working on tracking down where in the source code they have the upload file
-
https://jupyterlab.readthedocs.io/en/stable/api/
- Jupyterlite codebase uses Jupyterlab API for a lot of its functionality
-
https://github.com/jupyterlab/jupyterlab/blob/7d00c92aa951ad417f220460cd06c7237ebcc9d2/packages/filebrowser/style/base.css
- CSS file used for "Upload File"
-
https://github.com/jupyterlab/jupyterlab/tree/7d00c92aa951ad417f220460cd06c7237ebcc9d2/packages/filebrowser
- "A JupyterLab package which provides a file browser to the application. Document operations (such as opening, closing, renaming and moving), are delegated to the document manager"
-
https://github.com/jupyterlab/jupyterlab/tree/7d00c92aa951ad417f220460cd06c7237ebcc9d2/packages/docmanager
- "A JupyterLab package which handles interactions with documents and the file system. It opens and closes files, creates new views onto files, and keeps track of open documents."
-
https://github.com/jupyterlab/jupyterlab/tree/7d00c92aa951ad417f220460cd06c7237ebcc9d2/packages/filebrowser/src Contains the files for uploading (upload.ts, uploadStatus.tsx, model.ts...)
-
https://github.com/jupyterlab/jupyterlab/blob/7d00c92aa951ad417f220460cd06c7237ebcc9d2/packages/docmanager/src/tokens.ts Contains function to overwrite a file
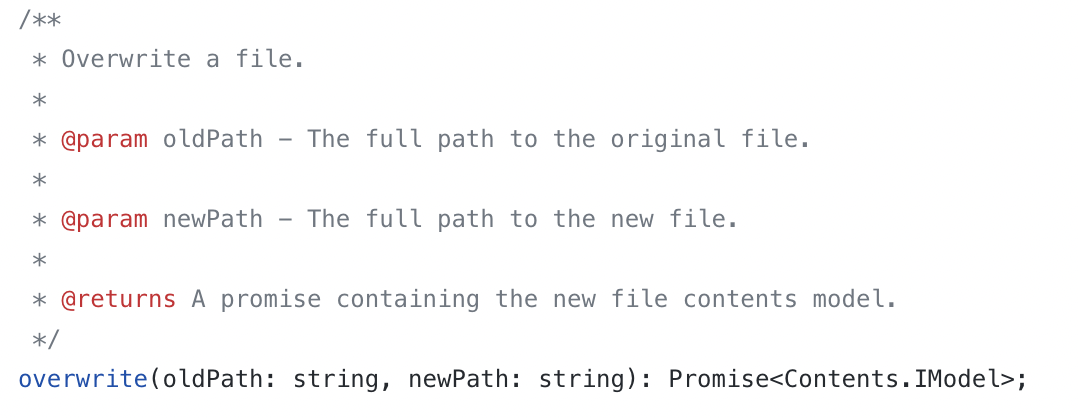
-
Like I said previously, jupyterlite relies heavily on jupyterlab's API. This is where all of the file editing options are. My next task is to try and figure out how we can attach a blob or use the overwrite function that they have already written to somehow overwrite or swap files
@pdxjohnny check this out when you have a second. Added lots of stuff. Want to touch base on next steps. I'll keep working on this and figuring out how we could accomplish this using what we know now
- https://github.com/jupyterlite/jupyterlite/issues/430 and https://github.com/jupyterlab/jupyterlab/pull/11387
- Issues from Jupyterlite Github and Jupyterlabs Github. Goes through handling different types of file uploads (notebook, csv, opening from url). Could be useful to us in the future when trying to trigger the file upload mechanism and how it handles different types of files, especially notebooks
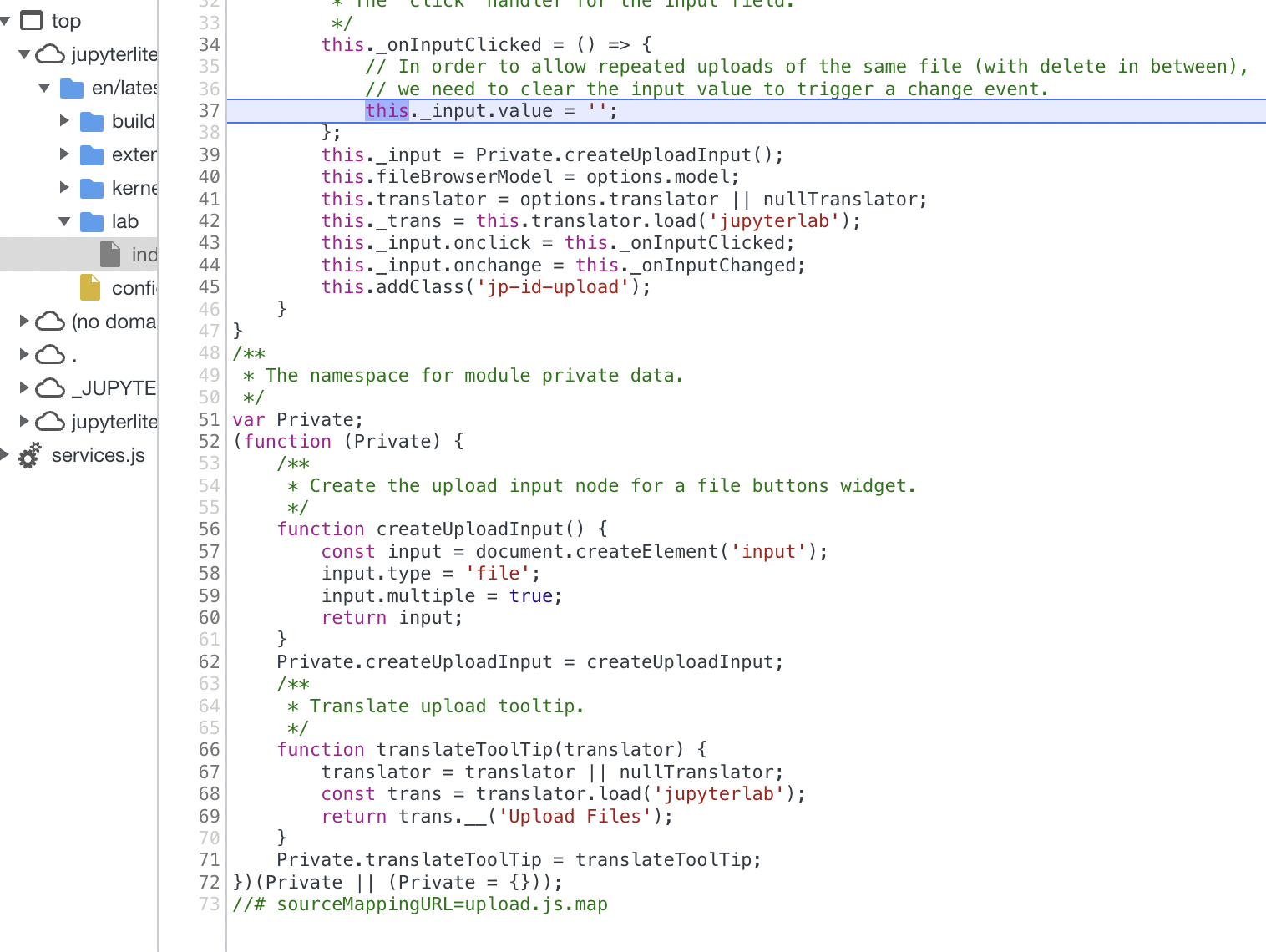

-
Working with devtools to trigger upload file mechanism. This is what I have to target somehow when inserting "the blob." This is the break down of when the onClick event listener is triggered on the file upload button
-
Event listener (onClick) is trigger on
div.lm-Widget p-Widget jp-ToolbarButton jp-id-upload jp-Toolbar-item

- This is the closest I have gotten to triggering onClick event. Whenever I try to do
.click()after grabbing the element, it comes back as undefined. But when I use the dispatchEvent, it comes back astrue
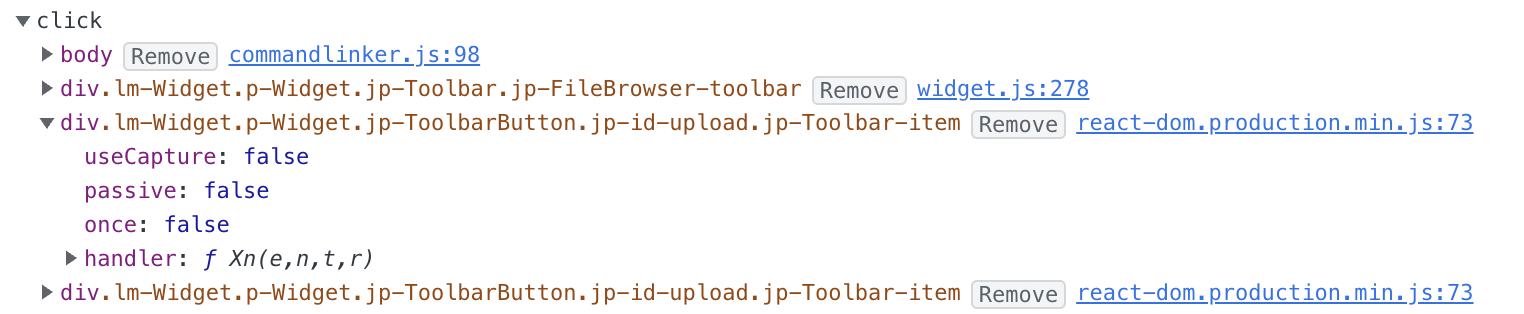
- Click event for
div.lm-Widget p-Widget jp-ToolbarButton jp-id-upload jp-Toolbar-itemlisted right here in the console. I know it's there.
- React actually captures mouse up/down rather than click.
Source: https://stackoverflow.com/questions/40091000/simulate-click-event-on-react-element
const mouseClickEvents = ['mousedown', 'click', 'mouseup'];
function simulateMouseClick(element){
mouseClickEvents.forEach(mouseEventType =>
element.dispatchEvent(
new MouseEvent(mouseEventType, {
view: window,
bubbles: true,
cancelable: true,
buttons: 1
})
)
);
}
var element = document.querySelector('div[class="UFIInputContainer"]');
simulateMouseClick(element);
- it then brings up a file dialog
Look for fileBrowserModel in Scopes, look for it's methods within the console window: https://github.com/jupyterlab/jupyterlab/blob/7dc7b9f1a30f75dadef9131d5fc6e1aa2160769a/packages/filebrowser/src/model.ts#L61
https://github.com/jupyterlab/jupyterlab/blob/7d00c92aa951ad417f220460cd06c7237ebcc9d2/packages/filebrowser/src/upload.ts#L24-L26
Is is a shorthand for a context local scoping, it binds the method to this.
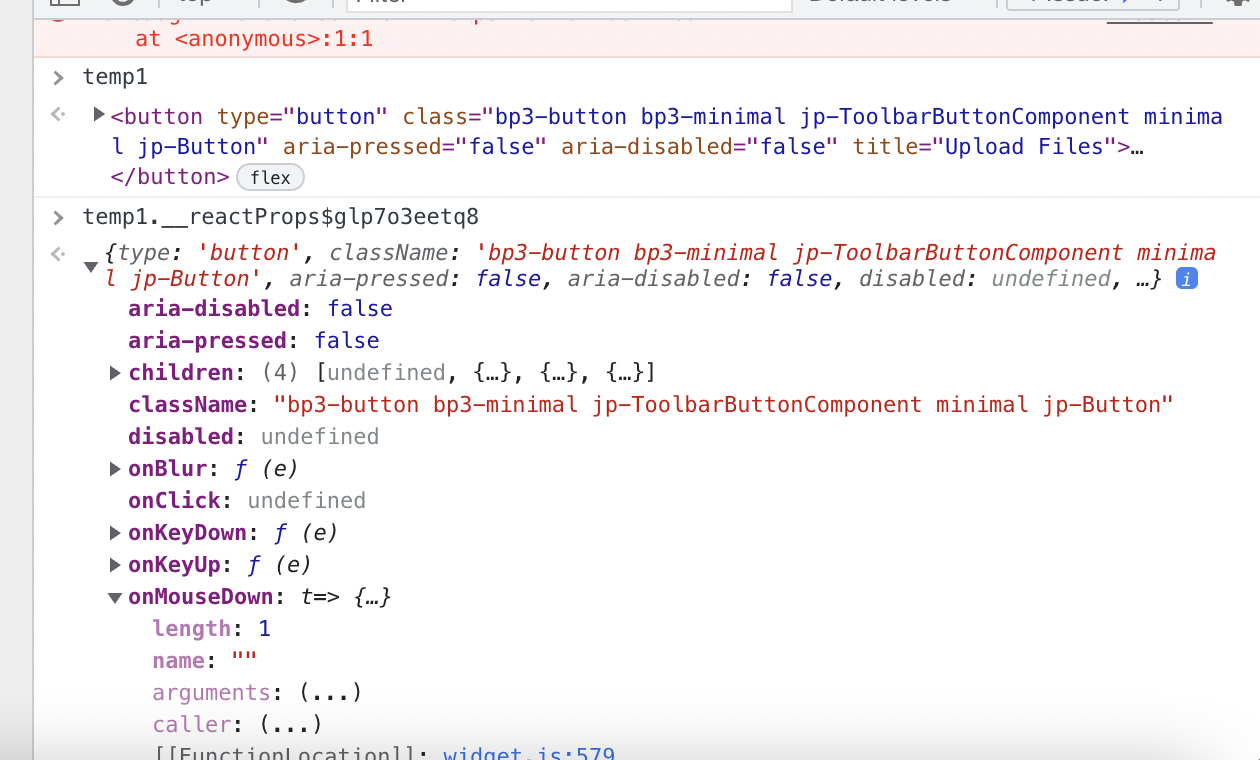
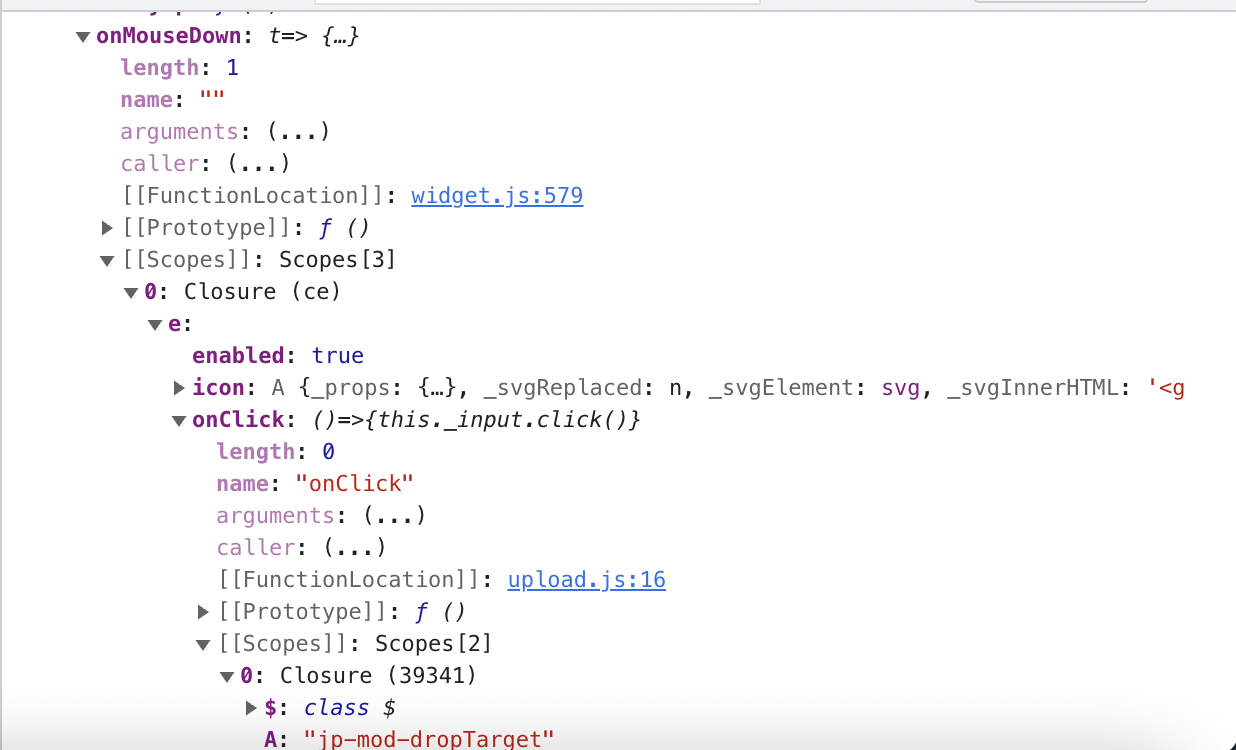
Working on finding _input
Set the button to a global variable then did temp1._reactProps and looking through that to try to find ._input
We think we found and object within the closure scope which has access to the kernel manager
https://github.com/jupyterlab/jupyterlab/blob/17cf5fcd5caf563a55b811e5df6377db612f6cd3/packages/services/src/manager.ts#L30-L73
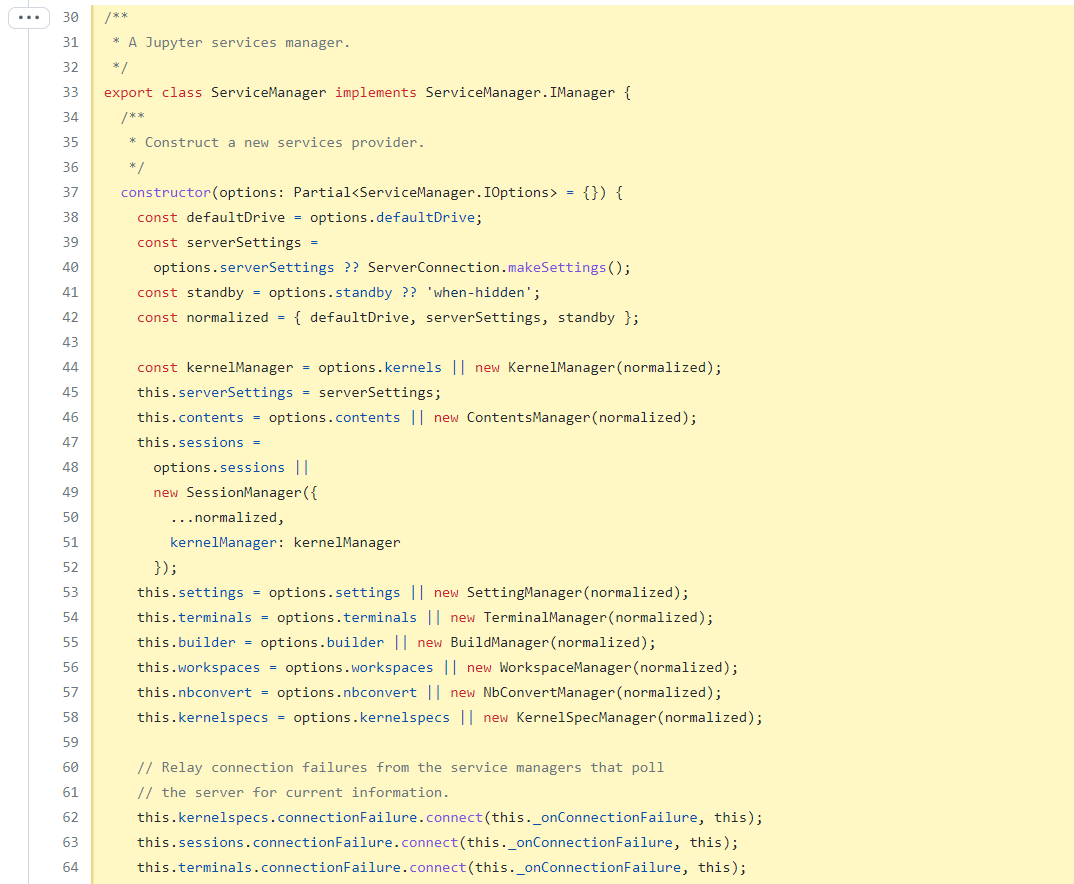
https://github.com/jupyterlab/jupyterlab/blob/7dc7b9f1a30f75dadef9131d5fc6e1aa2160769a/packages/filebrowser/src/model.ts#L409-L446
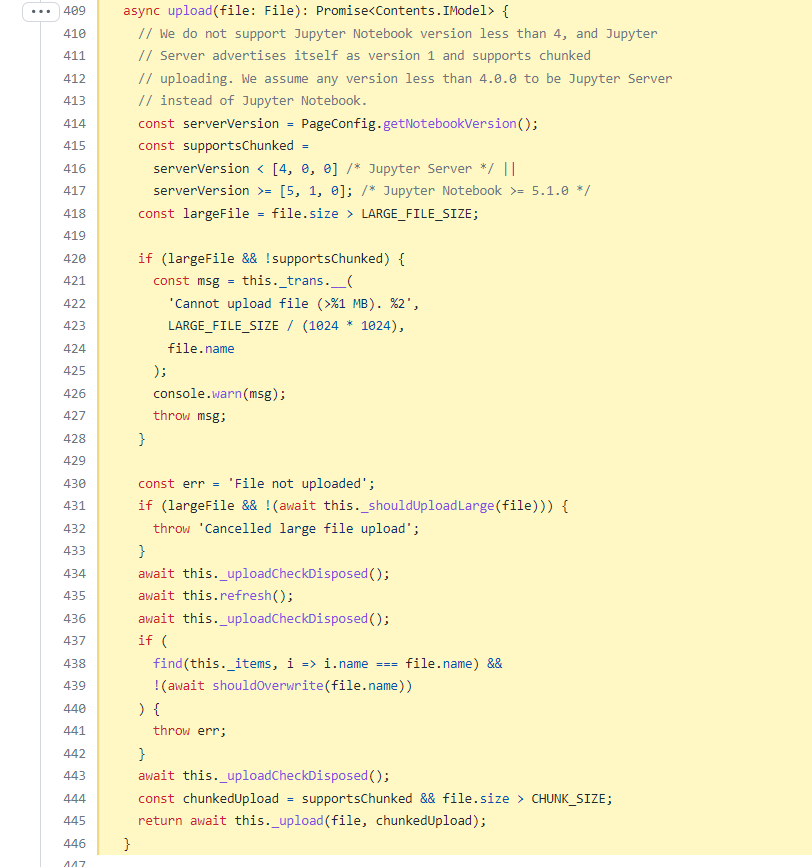
https://github.com/jupyterlab/jupyterlab/blob/7dc7b9f1a30f75dadef9131d5fc6e1aa2160769a/packages/filebrowser/src/model.ts#L477-L502

ServiceManager is probably this.manager.services, no it's not an instantiated instance.
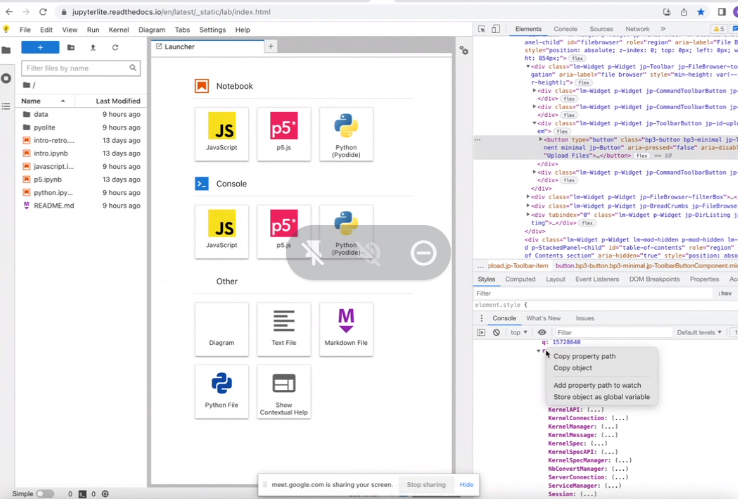
Saw getOpenFiles()
https://github.com/jupyterlab/jupyterlab/blob/b57ec962a7b8cda65bb89a72223f195564f8387a/packages/filebrowser/src/opendialog.ts
https://github.com/reactjs/reactjs.org/issues/3896
https://developer.mozilla.org/en-US/docs/Web/JavaScript/Reference/Global_Objects/Function/bind
Looking for '[[BoundThis]]'

Working on spinning up my own instance and trouble shooting the 404 error

I'm able to get my own instance of the jupyterlite-demo running with Github Pages at https://clairefollett.github.io/jupyterlite_demo/lab/index.html
https://jupyterlite.readthedocs.io/en/latest/quickstart/standalone.html - Deploying Jupyterlite on a standalone server or locally

Finally got it running with yarn serve:py and cd app && python -m http.server -b 127.0.0.1 8000
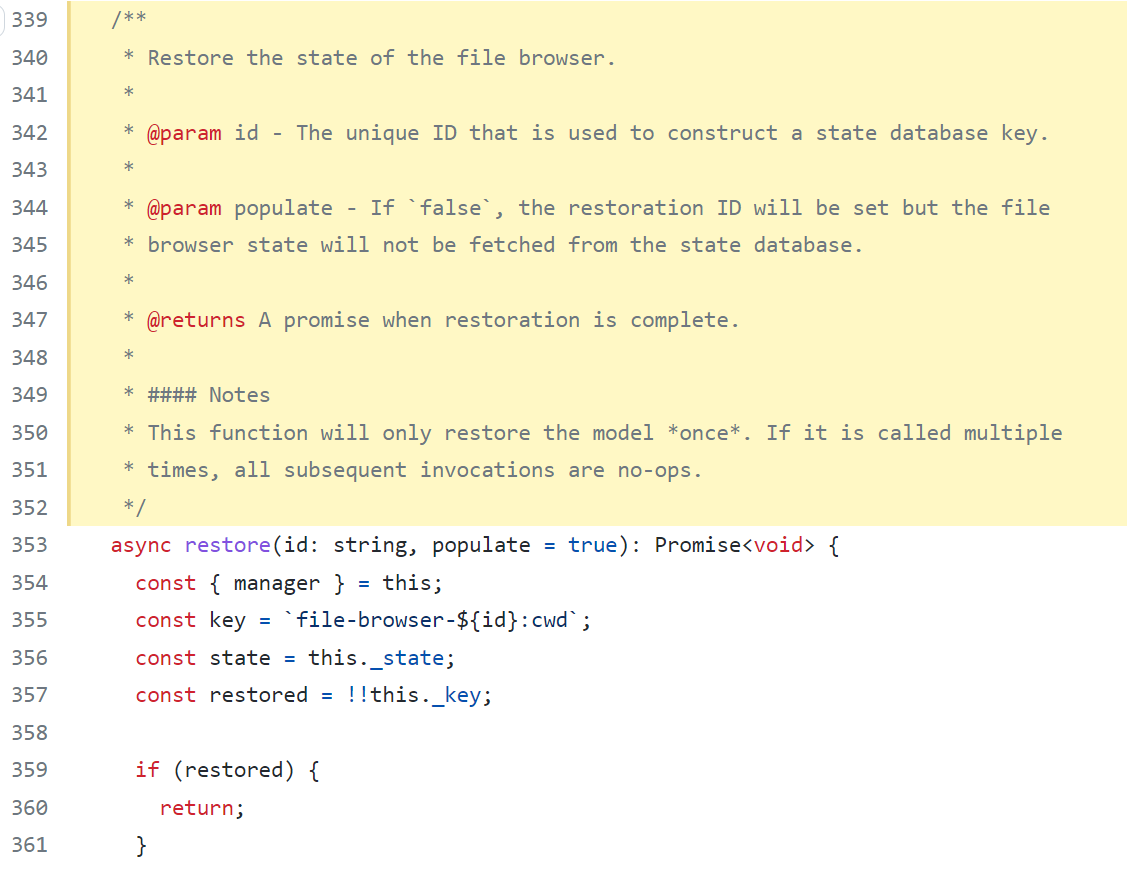
The constructor where this._input is in the filebrowser under Jupyterlab/packages/filebroswer/src/index.ts Jupyterlite uses Jupyterlab API and uses the filebrowser package. Next steps are trying to figure out how to grab this._input from the API and edit it I've been mainly searching through files and playing around with it - I know there's a way


jupyterlite/packages/application-extension/src/index.tsx - this file is importing objects from the fileBrowser module, which could be a good place to try and edit this._input
- Claire trying to
- References
- https://jupyterlite.readthedocs.io/en/latest/howto/configure/rtc.html
- https://jupyterlite.readthedocs.io/en/latest/howto/content/share.html
?path=filename.py
- https://github.com/jupyterlab/jupyterlab/blob/5c04fa6e8f8996608fa8d36be071d8d17a85a061/packages/filebrowser/src/model.ts#L339-L352
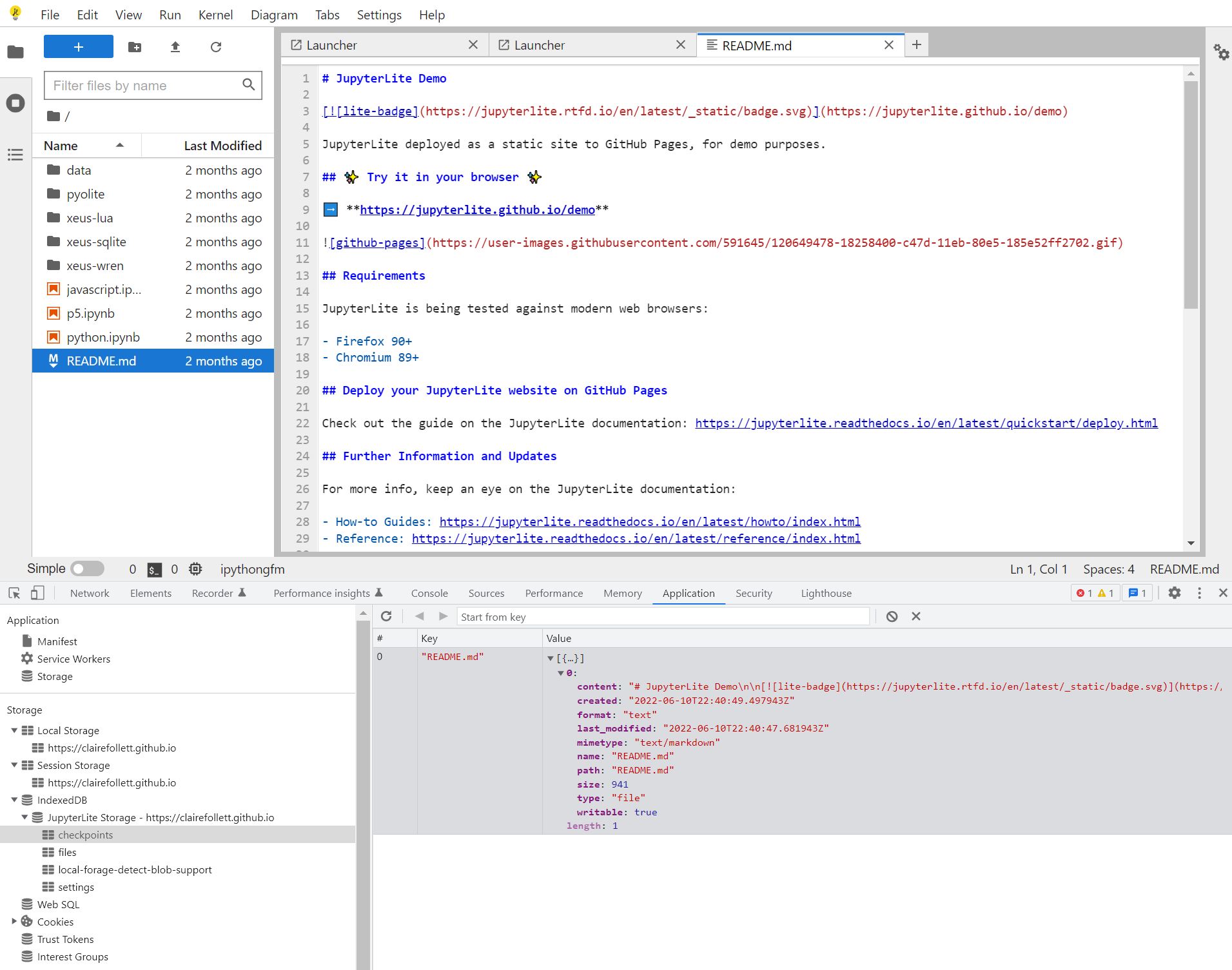
-
Can we edit content within checkpoints within IndexDB directly? Will it load on
refresh()? -
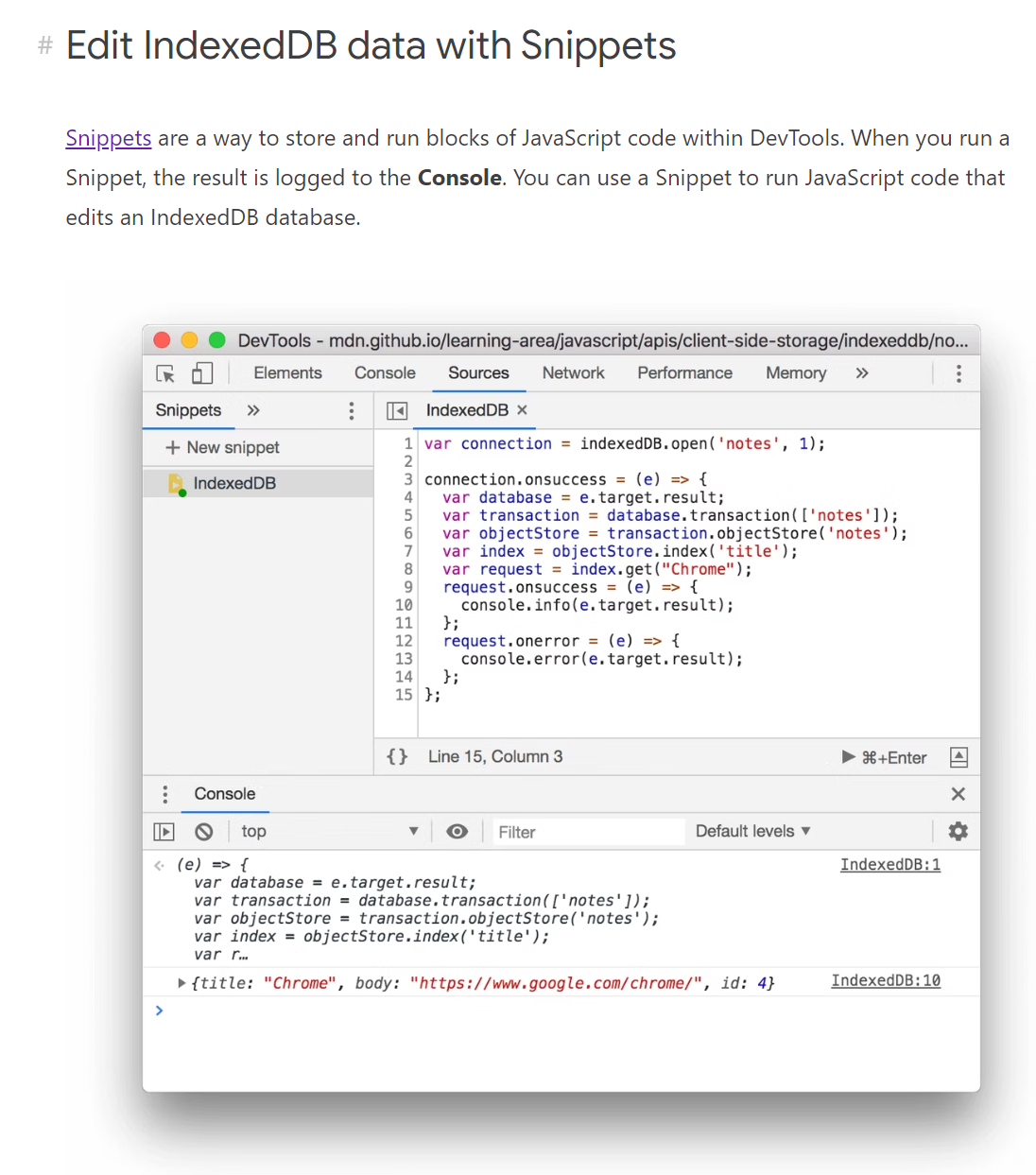
TIL: You can add snippets to chrome
- https://developer.chrome.com/docs/devtools/storage/indexeddb/#snippets
VM400 IndexedDB:17
Event {isTrusted: true, type: 'error', target: IDBOpenDBRequest, currentTarget: IDBOpenDBRequest, eventPhase: 2, …}
isTrusted: true
bubbles: true
cancelBubble: false
cancelable: true
composed: false
currentTarget: null
defaultPrevented: false
eventPhase: 0
path: []
returnValue: true
srcElement: IDBOpenDBRequest {onblocked: null, onupgradeneeded: null, result: undefined, error: DOMException: The requested version (1) is less than the existing version (4)., source: null, …}
target: IDBOpenDBRequest {onblocked: null, onupgradeneeded: null, result: undefined, error: DOMException: The requested version (1) is less than the existing version (4)., source: null, …}
timeStamp: 875442.200000003
type: "error"
[[Prototype]]: Event
var connection = indexedDB.open('JupyterLite Storage', 4)
connection.onsuccess = (e) => {
var database = e.target.result;
console.log(database)
var transaction = database.transaction(['checkpoints']);
var objectStore = transaction.objectStore('checkpoints');
console.log(objectStore)
var index = objectStore.index('Key');
var request = index.get("README.md")
request.onsuccess = (e) => {
console.info(e.target.result)
}
request.onerror = (e) => {
console.error(e.target.result)
}
}
connection.onerror = (e) => {
console.error(e)
}
- Console logs:
IDBDatabase {name: 'JupyterLite Storage', version: 4, objectStoreNames: DOMStringList, onabort: null, onclose: null, …}name: "JupyterLite Storage"objectStoreNames: DOMStringList0: "checkpoints"1: "files"2: "local-forage-detect-blob-support"3: "settings"length: 4[[Prototype]]: DOMStringListonabort: nullonclose: nullonerror: nullonversionchange: nullversion: 4[[Prototype]]: IDBDatabase
IndexedDB:8 IDBObjectStore {name: 'checkpoints', keyPath: null, indexNames: DOMStringList, transaction: IDBTransaction, autoIncrement: false}
- If we populate the files ObjectStore within the IndexDB, and provide the path via
?path=then it will load the document.- https://developer.mozilla.org/en-US/docs/Web/API/IDBObjectStore/get
- https://developer.mozilla.org/en-US/docs/Web/API/IDBFactory/open
- https://intel.github.io/dffml/main/examples/notebooks/moving_between_models.ipynb
- https://intel.github.io/dffml/main/examples/notebooks/moving_between_models.html
{
"name": "AliceisHere.drawio.xml",
"path": "AliceisHere.drawio.xml",
"last_modified": "2022-07-26T23:43:46.141Z",
"created": "2022-07-26T23:43:46.105Z",
"format": "text",
"mimetype": "application/json",
"content": "<mxfile host=\"app.diagrams.net\" modified=\"2022-07-20T22:07:58.641Z\" agent=\"5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/103.0.0.0 Safari/537.36\" etag=\"x_QOLTbdd3LFU5UQZKz1\" version=\"20.1.3\" type=\"device\"><diagram id=\"XeFC43bqpgZcxJIsIYqh\" name=\"Page-1\">jZPBboMwDIafhmOnljAKx5aVdodphx4q7YaIC9ECYSFt6Z5+TnEKrJq0CySfHdux/3gsqbqtzpryTXGQnj/nncdePN9nYYBfC649WCyee1BowQkNYC++geCc6ElwaCeORilpRDOFuapryM2EZVqry9TtqOQ0a5MV8AD2eSYf6UFwU9K12HLgOxBF6TJHZKgy50sXacuMq8sIsY3HEq2U6VdVl4C0rXNt6c+lf1jvdWmozX8OhOkO5OZg4vjIovA9lpuP1xlFOWfyRPf1/FBivPVRYVis2lypE+HXSTnDrL3NaYUOQdTgqNeDHVeF/a+kyMHmb/GzAw0uMpbYB+/9qDv3PL6BzvLWaPUJiZJKI69VDTa5kPIXKk0lcbeg0khBQeT2FNjaz6CNwNFiaUWNrBKcW+M6I5BjL0Fb0Da9mI6iA06hXGLPZ2kax2l6r348BddSzAXdCNFUtqAqMPqKLmRdxqQQeiFByPr9ZdDbYk6vphxpLXhiEQmdRF7cgw9CwAVpwW0Hzd1so3fLNj8=</diagram></mxfile>",
"size": 75,
"writable": true,
"type": "file"
}
- https://github.com/intel/dffml/blob/748c1b9072a1a2f4f8583e6f432792ad5cb17857/docs/conf.py#L86-L99
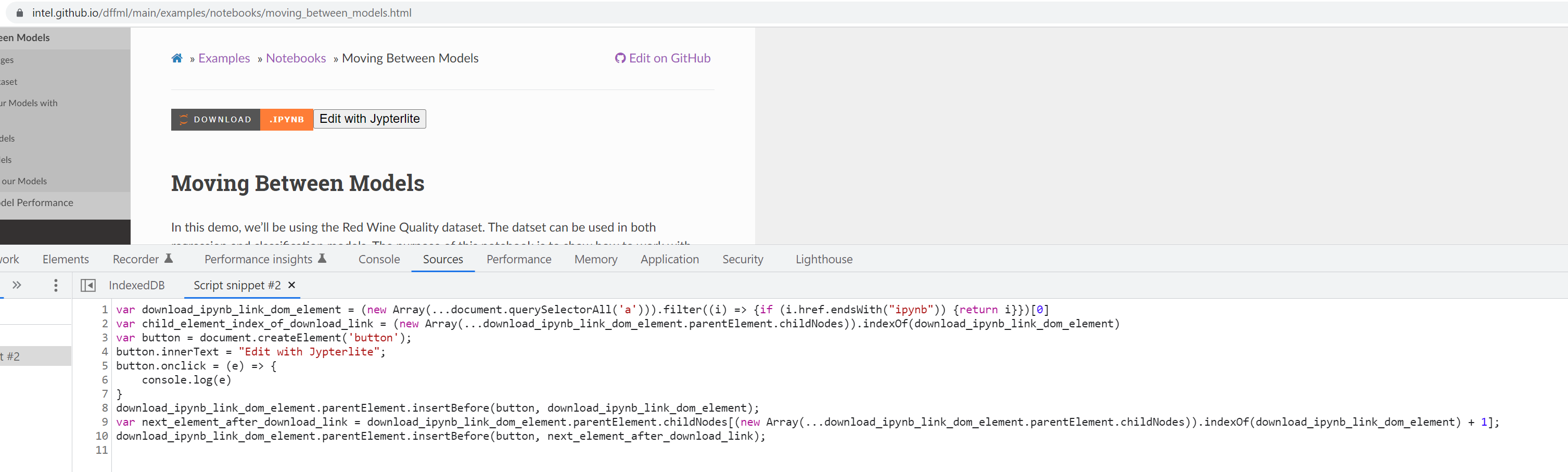
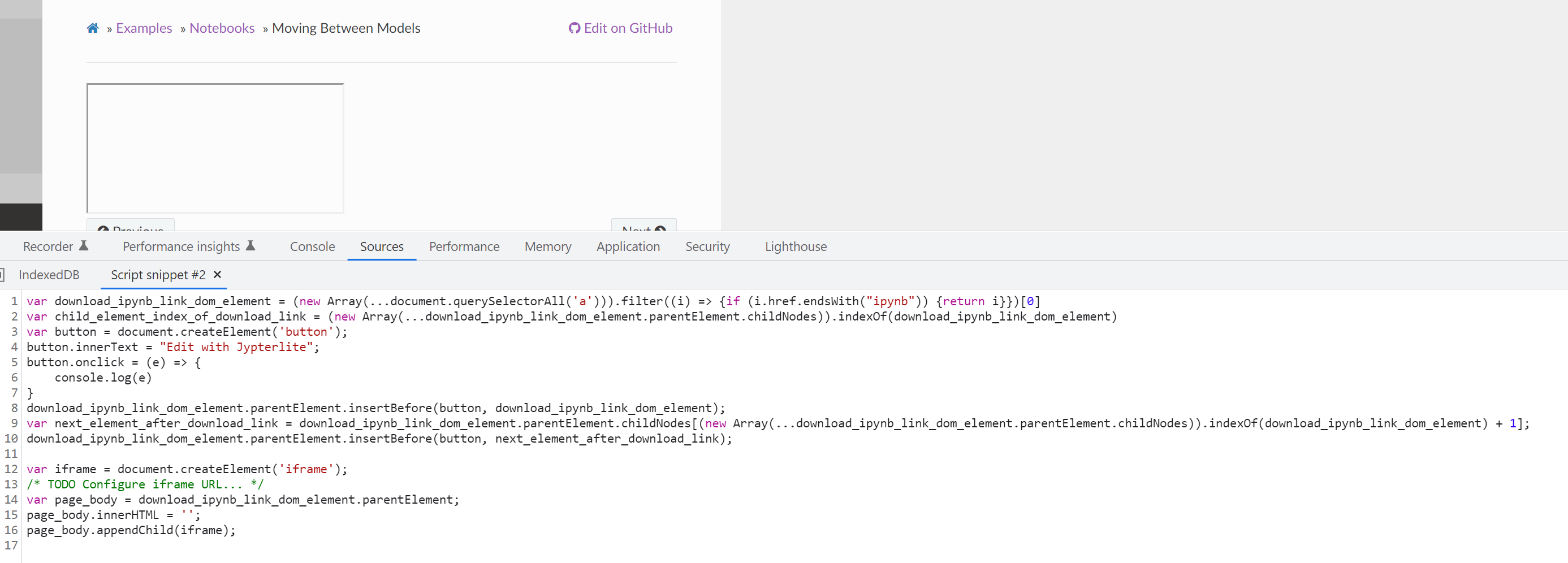
- Now we need to add the snippet script on notebook page load and trigger on click
- Add a another button for
Live Edit?
https://github.com/intel/dffml/blob/748c1b9072a1a2f4f8583e6f432792ad5cb17857/docs/conf.py#L117-L118
https://github.com/intel/dffml/blob/b7594381fd5f409a8ff20707d4fb836cc31174fb/dffml/service/dev.py#L1084-L1090
- We will put the JavaScript snippet we have in the docs using
add_js_file() - We only want to run on pages that have links to
.ipynbfiles. - If we detect a link, we should add a button to the
- Need to figure out how to create
objectStoreif not exists. - Need to figure out how to trigger load of jypterlite with
?path=filename.extension- Consider using an
iframe- Then you can set the path and you can just
document.appendChildthe
- Then you can set the path and you can just
- Consider using an
- Combine the following with db insertion code into a file which you
add_js_file()during docs build.
var download_ipynb_link_dom_element = (new Array(...document.querySelectorAll('a'))).filter((i) => {if (i.href.endsWith("ipynb")) {return i}})[0]
var child_element_index_of_download_link = (new Array(...download_ipynb_link_dom_element.parentElement.childNodes)).indexOf(download_ipynb_link_dom_element)
var button = document.createElement('button');
button.innerText = "Edit with Jypterlite";
download_ipynb_link_dom_element.parentElement.insertBefore(button, download_ipynb_link_dom_element);
(new
var next_element_after_download_link = download_ipynb_link_dom_element.parentElement.childNodes[(new Array(...download_ipynb_link_dom_element.parentElement.childNodes)).indexOf(download_ipynb_link_dom_element) + 1];
undefined
download_ipynb_link_dom_element.parentElement.insertBefore(button, next_element_after_download_link);
# To hotswap
var iframe = document.createElement('iframe');
# TODO Configure iframe URL...
var page_body = download_ipynb_link_dom_element.parentElement;
page_body.innerHTML = '';
page_body.appendChild(iframe);
var connection = indexedDB.open('JupyterLite Storage');
connection.onsuccess = (e) => {
var database = e.target.result;
var transaction = database.transaction(['files'], 'readwrite');
var objectStore = transaction.objectStore('files');
var request = objectStore.getAllKeys();
request.onsuccess = (e) => {
console.info(e.target.result);
var request = objectStore.get(e.target.result[0]);
request.onsuccess = (e) => {
console.info(e.target.result);
};
request.onerror = (e) => {
console.error(e.target.result);
};
};
request.onerror = (e) => {
console.error(e.target.result);
};
};
connection.onerror = (e) => {
console.error(e);
};
The snippet that allows us to grab things from local storage
https://developer.mozilla.org/en-US/docs/Web/API/IDBObjectStore https://developer.mozilla.org/en-US/docs/Web/API/IDBObjectStore/get
var connection = indexedDB.open('JupyterLite Storage');
var file = {
"name": "moving_between_models.ipynb",
"path": "moving_between_models.ipynb",
"last_modified": "2022-07-26T23:45:32.537Z",
"created": "2022-07-26T23:45:32.522Z",
"format": "json",
"mimetype": "application/json",
"content": {
"cells": [
{
"cell_type": "markdown",
"metadata": {},
"source": [
"# Moving Between Models\n",
"\n",
"In this demo, we'll be using the Red Wine Quality dataset. The datset can be used in both regression and classification models. The purpose of this notebook is to show how to work with multiple models in DFFML."
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Import Packages\n",
"\n",
"Let us import dffml and other packages that we might need."
]
},
{
"cell_type": "code",
"execution_count": 1,
"metadata": {},
"outputs": [],
"source": [
"from dffml import *"
]
},
{
"cell_type": "code",
"execution_count": 2,
"metadata": {},
"outputs": [],
"source": [
"import asyncio\n",
"import nest_asyncio"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"To use asyncio in a notebook, we need to use nest_asycio.apply()"
]
},
{
"cell_type": "code",
"execution_count": 3,
"metadata": {},
"outputs": [],
"source": [
"nest_asyncio.apply()"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Build our Dataset\n",
"\n",
"Dffml has a very convinient function `cached_download()` that can be used to download datasets and make sure you don't download them if you have already."
]
},
{
"cell_type": "code",
"execution_count": 4,
"metadata": {},
"outputs": [],
"source": [
"data_path = await cached_download(\n",
" \"https://archive.ics.uci.edu/ml/machine-learning-databases/wine-quality/winequality-red.csv\",\n",
" \"wine_quality.csv\",\n",
" \"789e98688f9ff18d4bae35afb71b006116ec9c529c1b21563fdaf5e785aea8b3937a55a4919c91ca2b0acb671300072c\",\n",
")"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"In Dffml, we try to use asynchronicity where we can, to get that extra bit of performance. Let's use the async version of `load()` to load the dataset that we just downloaded into a source. We can easily achieve this by declaring a `CSVSource` with the data_path and the delimiter since the data we downloaded seems to have a non-comma delimiter. \n",
"\n",
"After that, we can just create an array of `records` by loading each one through the `load()` function.\n",
"\n",
"Feel free to also try out the no async version of `load()`."
]
},
{
"cell_type": "code",
"execution_count": 5,
"metadata": {},
"outputs": [],
"source": [
"async def load_dataset(data_path):\n",
" data_source = CSVSource(filename=data_path, delimiter=\";\")\n",
" data = [record async for record in load(data_source)]\n",
" return data\n",
"\n",
"\n",
"data = asyncio.run(load_dataset(data_path))"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Dffml lets you visualize a record in quite a neat fashion. Lets have a look."
]
},
{
"cell_type": "code",
"execution_count": 6,
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"\n",
"\tKey:\t0\n",
" Record Features\n",
"+----------------------------------------------------------------------+\n",
"| fixed acidity | 7.4 |\n",
"+----------------------------------------------------------------------+\n",
"| volatile acidity| 0.7 |\n",
"+----------------------------------------------------------------------+\n",
"| citric acid | 0 |\n",
"+----------------------------------------------------------------------+\n",
"| residual sugar | 1.9 |\n",
"+----------------------------------------------------------------------+\n",
"| chlorides | 0.076 |\n",
"+----------------------------------------------------------------------+\n",
"|free sulfur dioxi| 11 |\n",
"+----------------------------------------------------------------------+\n",
"|total sulfur diox| 34 |\n",
"+----------------------------------------------------------------------+\n",
"| density | 0.9978 |\n",
"+----------------------------------------------------------------------+\n",
"| pH | 3.51 |\n",
"+----------------------------------------------------------------------+\n",
"| sulphates | 0.56 |\n",
"+----------------------------------------------------------------------+\n",
"| alcohol | 9.4 |\n",
"+----------------------------------------------------------------------+\n",
"| quality | 5 |\n",
"+----------------------------------------------------------------------+\n",
" Prediction: Undetermined \n",
"\n",
"1599\n"
]
}
],
"source": [
"print(data[0], \"\\n\")\n",
"print(len(data))"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"Lets split our dataset into train and test splits."
]
},
{
"cell_type": "code",
"execution_count": 7,
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"1599 1279 320\n"
]
}
],
"source": [
"train_data = data[320:]\n",
"test_data = data[:320]\n",
"print(len(data), len(train_data), len(test_data))"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Instantiate our Models with parameters\n",
"Dffml makes it quite easy to load multiple models dynamically using the `Model.load()` function. After that, you just have to parameterize the loaded models and they are ready to train interchangably!\n",
"\n",
"For this example, we'll be demonstrating 2 models but you can feel free to try more than 2 models in a similar fashion."
]
},
{
"cell_type": "code",
"execution_count": 8,
"metadata": {},
"outputs": [],
"source": [
"ScikitLORModel = Model.load(\"scikitlor\")\n",
"ScikitETCModel = Model.load(\"scikitetc\")\n",
"\n",
"features = Features(\n",
" Feature(\"fixed acidity\", int, 1),\n",
" Feature(\"volatile acidity\", int, 1),\n",
" Feature(\"citric acid\", int, 1),\n",
" Feature(\"residual sugar\", int, 1),\n",
" Feature(\"chlorides\", int, 1),\n",
" Feature(\"free sulfur dioxide\", int, 1),\n",
" Feature(\"total sulfur dioxide\", int, 1),\n",
" Feature(\"density\", int, 1),\n",
" Feature(\"pH\", int, 1),\n",
" Feature(\"sulphates\", int, 1),\n",
" Feature(\"alcohol\", int, 1),\n",
")\n",
"\n",
"predict_feature = Feature(\"quality\", int, 1)\n",
"\n",
"model1 = ScikitLORModel(\n",
" features=features,\n",
" predict=predict_feature,\n",
" location=\"scikitlor\",\n",
" max_iter=150,\n",
")\n",
"model2 = ScikitETCModel(\n",
" features=features,\n",
" predict=predict_feature,\n",
" location=\"scikitetc\",\n",
" n_estimators=150,\n",
")"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Train our Models\n",
"Finally, our models are ready to be trained using the `high-level` API. Let's make sure to pass each record as a parameter by simply using the unpacking operator(*)."
]
},
{
"cell_type": "code",
"execution_count": 9,
"metadata": {},
"outputs": [],
"source": [
"await train(model1, *train_data)\n",
"await train(model2, *train_data)"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Test our Models\n",
"To test our model, we'll use the `score()` function in the `high-level` API.\n",
"\n",
"We ask for the accuracy to be assessed using the Mean Squared Error method."
]
},
{
"cell_type": "code",
"execution_count": 10,
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Accuracy1: 0.4625\n",
"Accuracy2: 0.46875\n"
]
}
],
"source": [
"MeanSquaredErrorAccuracy = AccuracyScorer.load(\"mse\")\n",
"\n",
"scorer = MeanSquaredErrorAccuracy()\n",
"\n",
"print(\"Accuracy1:\", await score(model1, scorer, predict_feature, *test_data))\n",
"print(\"Accuracy2:\", await score(model2, scorer, predict_feature, *test_data))"
]
},
{
"cell_type": "markdown",
"metadata": {},
"source": [
"### Predict using our Models\n",
"Let's make predictions and see what they look like for each model using the `predict` function in the `high-level` API.\n",
"\n",
"Note that the `predict` function returns an asynciterator of a `Record` Object that contains a tuple of record.key, features and predictions.\n",
"\n",
"For the sake of visualizing data, we'll keep the predictions to a few records."
]
},
{
"cell_type": "code",
"execution_count": 11,
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"{'confidence': nan, 'value': 5}\n",
"{'confidence': nan, 'value': 5}\n",
"{'confidence': nan, 'value': 5}\n",
"{'confidence': nan, 'value': 5}\n",
"{'confidence': nan, 'value': 5}\n"
]
}
],
"source": [
"# Modified Test_data\n",
"m_test_data = test_data[:5]\n",
"# Predict and view Predictions for model 1\n",
"async for i, features, prediction in predict(model1, *m_test_data):\n",
" features[\"quality\"] = prediction[\"quality\"]\n",
" print(features[\"quality\"])"
]
},
{
"cell_type": "code",
"execution_count": 12,
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"{'confidence': nan, 'value': 5}\n",
"{'confidence': nan, 'value': 5}\n",
"{'confidence': nan, 'value': 5}\n",
"{'confidence': nan, 'value': 6}\n",
"{'confidence': nan, 'value': 5}\n"
]
}
],
"source": [
"# Predict and view Predictions for model 2\n",
"async for i, features, prediction in predict(model2, *m_test_data):\n",
" features[\"quality\"] = prediction[\"quality\"]\n",
" print(features[\"quality\"])"
]
}
],
"metadata": {
"celltoolbar": "Tags",
"kernelspec": {
"display_name": "Python 3 (ipykernel)",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.8.10"
}
},
"nbformat": 4,
"nbformat_minor": 4
},
"size": 75,
"writable": true,
"type": "notebook"
}
connection.onsuccess = (e) => {
var database = e.target.result;
var transaction = database.transaction(['files'], 'readwrite');
var objectStore = transaction.objectStore('files');
var request = objectStore.clear();
request.onsuccess = (e) => {
console.info(e.target.result);
var request = objectStore.add(file, 'test.ipynb');
request.onsuccess = (e) => {
console.info(e.target.result);
};
request.onerror = (e) => {
console.error(e.target.result);
};
};
request.onerror = (e) => {
console.error(e.target.result);
};
};
connection.onerror = (e) => {
console.error(e);
};
NOTE `cat command should be ^C'd after paste
$ cat > patchfile
Paste the following
diff --git a/model/scratch/dffml_model_scratch/anomaly_detection_scorer.py b/model/scratch/dffml_model_scratch/anomaly_detection_scorer.py
index 0c289f575..54138de42 100644
--- a/model/scratch/dffml_model_scratch/anomaly_detection_scorer.py
+++ b/model/scratch/dffml_model_scratch/anomaly_detection_scorer.py
@@ -11,7 +11,7 @@ from dffml.accuracy import (
AccuracyContext,
)
-from model.scratch.dffml_model_scratch.anomalydetection import (
+from .anomalydetection import (
getF1,
multivariateGaussian,
findIndices,
diff --git a/scripts/docs.py b/scripts/docs.py
index 77e5391e2..f54c7cb4f 100644
--- a/scripts/docs.py
+++ b/scripts/docs.py
@@ -167,7 +167,10 @@ def gen_docs(entrypoint: str, maintenance: str = "Official"):
# Skip duplicates
if i.name in done:
continue
- cls = i.load()
+ try:
+ cls = i.load()
+ except:
+ continue
plugin_type = "_".join(cls.ENTRY_POINT_NAME)
if plugin_type == "opimp":
plugin_type = "operation"
Ctrl-C
Then
$ patch -p1 < patchfile
For installing pandoc
$ mkdir -p ~/.local && curl -L https://github.com/jgm/pandoc/releases/download/2.19.2/pandoc-2.19.2-linux-amd64.tar.gz | tar -xvz -C ~/.local && export PATH="$PATH:$HOME/.local/bin" && mv ~/.local/pandoc-*/* ~/.local/
For building docs
$ dffml service dev docs -no-strict
We can put the JS under docs/_ext/
$ python -m http.server --directory pages/
We need to also build ensure a jypterlite static HTML build exists under pages/
- [ ] @Clairefollett to modify
dffml/serivce/dev.pyto run jypterlite static (follow jypterlite standalone quickstart) - [x] Confirm button adding code still works
- [x] Add basic button adding code to
docs/_ext/liveedit.js - [x] In
docs/conf.py, enable new JS viaadd_js_file(), file must be in_static/ - [x] On build in
dffml/service/dev.pywhen building docs, we should copy viashutil.copy()into the_staticdirectory - [x] JS running after DOM button load so we can watch it run
- https://developer.mozilla.org/en-US/docs/Web/API/Window/DOMContentLoaded_event
window.addEventListener('DOMContentLoaded', (event) => {
console.log('DOM fully loaded and parsed');
});
The console shows that the site is loading the file liveedit.js
Working on adding iframe. Came across this interesting article about embedding Jupyterlite REPL to existing websites: https://blog.jupyter.org/jupyter-everywhere-f8151c2cc6e8
May come in handy, may not
`
window.addEventListener('DOMContentLoaded', (event) => { console.log('DOM fully loaded and parsed');
var download_ipynb_link_dom_element = (new Array(...document.querySelectorAll('a'))).filter((i) => {if (i.href.endsWith("ipynb")) {return i}})[0];
var child_element_index_of_download_link = (new Array(...download_ipynb_link_dom_element.parentElement.childNodes)).indexOf(download_ipynb_link_dom_element);
var button = document.createElement('button');
button.innerText = "Edit with Jypterlite";
download_ipynb_link_dom_element.parentElement.insertBefore(button, download_ipynb_link_dom_element);
var next_element_after_download_link = download_ipynb_link_dom_element.parentElement.childNodes[(new Array(...download_ipynb_link_dom_element.parentElement.childNodes)).indexOf(download_ipynb_link_dom_element) + 1];
download_ipynb_link_dom_element.parentElement.insertBefore(button, next_element_after_download_link);
console.log("Success");
});
// To hotswap // var iframe = document.createElement('iframe'); // TODO Configure iframe URL... // var page_body = download_ipynb_link_dom_element.parentElement; // page_body.innerHTML = ''; // page_body.appendChild(iframe);`
liveedit_path = pages_path / "_static" / "liveedit.js" liveedit_source_path = pathlib.Path(__file__).parents[2].joinpath("docs", "_ext", "liveedit.js") shutil.copy(liveedit_source_path, liveedit_path)
dffml/service/dev.py
app.add_js_file("liveedit.js")
docs/conf.py
docs/_ext/liveedit.js