netcdf4-python
 netcdf4-python copied to clipboard
netcdf4-python copied to clipboard
Unexpectedly high memory usage opening netCDF4 file with many variables
Version : netCDF4-python 1.6.0 OS: Linux Python version: 3.9.15
I have a set of netCDF4 files that use substantially more memory to open than expected. I’ve included a reduced-size version of one of these files in a public repo here: https://github.com/dougiesquire/um_output_memory/blob/main/cj877a.pm000101_mon.1x1.nc4
That file is 1.5 MB on disk, but uses something like 20 MB of memory to open a single variable:
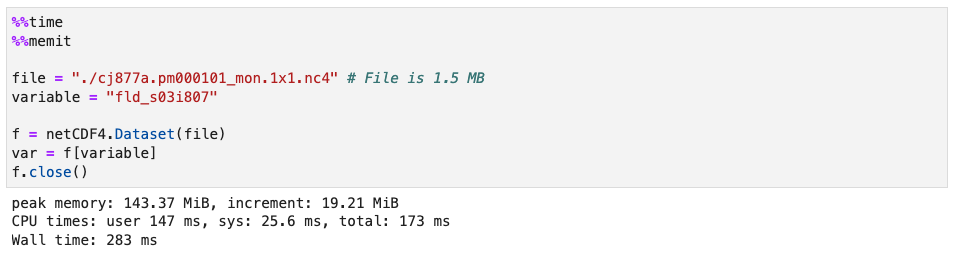
Because of this issue, I am unable to open and concatenate many such files.
I’d really appreciate any help understanding/debugging/fixing what the issue is here. In the repo linked above, there's also a notebook showing examples of the high memory usage when opening the reduced-size example file using netCDF4-python.
Some things to note
- Converting these files to NETCDF3 seems to fix the issue - the above code block with a NETCDF3 version of the same file uses ~1MB of memory.
- Interestingly, the memory footprint is essentially the same for the reduced-size files included in the above repo as for the original full-size files. The reduced-size files include only one spatial grid point, whereas the full size files include 27,648. It's almost like it's the metadata that is responsible for the large memory footprint…?
- These files contains 250 variables. I've never worked with NetCDF files containing this many variables - is the problem related to this perhaps?
- These files have
filling off. Out of desperation, I’ve tried recreating the data withfilling onbut that didn’t help. - Opening these files with
h5netcdfuses less memory, but takes a prohibitively long time.
netcdf4-python wraps the netcdf-c library, which in turn uses the HDF5 c library. I don't believe the large memory usage (which I was able to reproduce) is related to the python interface. Since you noted that using NETCDF3 fixes it, it's probably related to HDF5. I'm sorry but I don't have any suggestions for addressing this - perhaps you could get help on the netcdf-c issue tracker.
Thanks @jswhit, and thanks too for confirming you can reproduce the issue. I'll try to open something with netcdf-c as you suggest.