scadnano
 scadnano copied to clipboard
scadnano copied to clipboard
Find sequence
Implement 'Find' for a sequence: User hits Edit-> find (or keyboard shortcut: Cmd+F on Mac, Ctrl+F on other architectures), types a DNA sequence s, and if s is in the design it is clearly highlighted. Bonus: The find dialog could possibly have a tickbox (off by default) to find the complement of the sequence typed. Mega bonus: Find should work across nicks and crossovers (maybe another tickbox).
I created a doublon of that feature request, let me paste my message here instead:
A useful feature when working both from the source json .sc and the interface would be, in the interface, to be able to highlight a strand which has a given sequence.
Example usecase: in a design I receive from a colleague with a few hundres of staples, 2 of them are of length 31. I can easily get their sequence in the .sc file but really hard to figure out where they are exactly on the screen in the interface.
There's two options:
- Just search the substrings of DNA sequences of existing strands. (easy-ish)
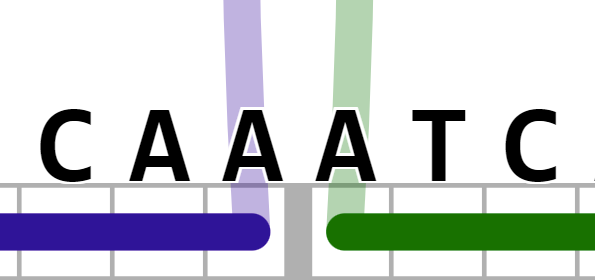
- Search for substrings "along a helix", i.e., searching for "CAAATC" highlight would highlight the following:
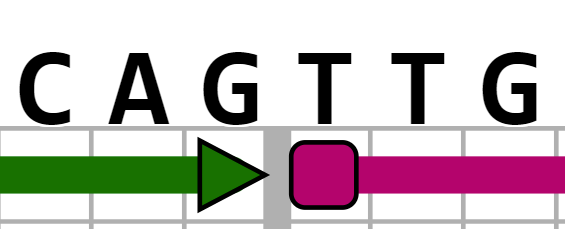
 and searching for "CAGTTG" would highlight this:
and searching for "CAGTTG" would highlight this: