RMG-database
 RMG-database copied to clipboard
RMG-database copied to clipboard
Forbidden H-Abs groups of multiple bond disprop cases
We currently forbid many H-abstraction reactions which are disproportionation, such as:
OH + C2H5 = H2O + birad-C2H4
But we don't forbid the similar cases involving multiple bonds:
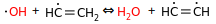
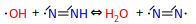 I think the above should be forbidden as well, and instead we should only keep the disprop equivalents:
I think the above should be forbidden as well, and instead we should only keep the disprop equivalents:
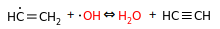
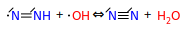
I think these are "simple" cases, and the fact that they aren't already forbidden in H-abs. makes me suspicious that perhaps I'm missing something. Any thoughts?
Hi,
Even after "make clean" and "make" process. I got similar error...
The file location
rajesh@pharos:~/Rajesh/New_jobs_after_Aug_11/MultiT_PM3_pdep_Prun_15000_multiple_concentration_single_presssure_conversion$
Error Message:
Created new reverse Alkene_to_1,2-birad reaction: C4H8O(4552) --> C4H8OJJ(5231)
Solving PDepNetwork #344 (propanal)
For reaction CH3CH2CO(355) + H(29) (included =true) propanal(368) (included =true)
Pressure-dependent rate coefficient at 679K 4.5Bar exceeds high-P-limit rate by factor of 2.4 .
Re-running fame with 501 grains.
Solving PDepNetwork #344 (propanal)
Solving PDepNetwork #583 (C14H29O2J)
ERROR: java.lang.NullPointerException
at jing.rxn.FastMasterEqn.readMeaningfulLine(FastMasterEqn.java:749)
at jing.rxn.FastMasterEqn.parseOutputStream(FastMasterEqn.java:854)
at jing.rxn.FastMasterEqn.runPDepCalculation(FastMasterEqn.java:383)
at jing.rxnSys.ReactionSystem.initializePDepNetwork(ReactionSystem.java:542)
at jing.rxnSys.ReactionModelGenerator.modelGeneration(ReactionModelGenerator.java:1472)
at RMG.main(RMG.java:96)
ERROR: java.lang.NullPointerException
at jing.rxnSys.Logger.log(Logger.java:161)
at jing.rxnSys.Logger.error(Logger.java:214)
at jing.rxn.FastMasterEqn.runPDepCalculation(FastMasterEqn.java:400)
at jing.rxnSys.ReactionSystem.initializePDepNetwork(ReactionSystem.java:542)
at jing.rxnSys.ReactionModelGenerator.modelGeneration(ReactionModelGenerator.java:1472)
at RMG.main(RMG.java:96)
Exception in thread "main" java.lang.NullPointerException
at jing.rxnSys.Logger.log(Logger.java:161)
at jing.rxnSys.Logger.critical(Logger.java:205)
at RMG.main(RMG.java:106)
To help interpret line numbers, note that the stack trace above was made with RMG at commit b688a393fe7c58442a717f49fdedf4ba5f884f6f
The NullPointerException is because it's trying to read more kinetics than there are in the fame output. Running fame.exe on the input file works without any error messages, but the output says it will contain 8856 phenomenological rate coefficients, but then reports only three.
The fame.log file ends as follows:
Fitting Chebyshev interpolation models...
Warning: One or more k(T,P) values for a net reaction was zero.
These have been set to 1e-300 to allow for k(T,P) interpolation model fitting.
c14h29o2j(547) -> c14h29o2j(1659)
0.0000000000000000 0.0000000000000000 0.0000000000000000
0.0000000000000000 0.0000000000000000 0.0000000000000000
0.0000000000000000 0.0000000000000000 0.0000000000000000
0.0000000000000000 0.0000000000000000 0.0000000000000000
0.0000000000000000 0.0000000000000000 0.0000000000000000
0.0000000000000000 0.0000000000000000 0.0000000000000000
I have made some extra commits to improve the error messages around this section of code, but this won't help to solve whatever the problem is in fame.exe, which perhaps @jwallen can look at next week. (I've a copy of 0583_4_input.txt in my home folder)
My commits should however allow it to fall back from ReservoirState to ModifiedStrongCollision method and try again, so I suggest you get my latest commits and restart @rajeshdparmar.
I seem to have a similar issue. Was going to post as new issue after discussion with @jwallen, but found this similar issue and figured I'd post it here:
Created new species: C10H8O2JJ(13834)
Created new BiradFromMultipleBond reaction: C10H8O2(13561) --> C10H8O2JJ(13834)
Ea raised by 0.7 from 7.8 to dHrxn(298K)=8.5 kcal/mol.
Solving PDepNetwork #9391 (C10H8O2JJ)
ERROR: java.lang.NullPointerException
at jing.rxn.FastMasterEqn.readMeaningfulLine(FastMasterEqn.java:745)
at jing.rxn.FastMasterEqn.parseOutputStream(FastMasterEqn.java:850)
at jing.rxn.FastMasterEqn.runPDepCalculation(FastMasterEqn.java:383)
at jing.rxnSys.ReactionSystem.initializePDepNetwork(ReactionSystem.java:542)
at jing.rxnSys.ReactionModelGenerator.modelGeneration(ReactionModelGenerator.java:1466)
at RMG.main(RMG.java:96)
ERROR: java.lang.NullPointerException
at jing.rxnSys.Logger.log(Logger.java:160)
at jing.rxnSys.Logger.error(Logger.java:213)
at jing.rxn.FastMasterEqn.runPDepCalculation(FastMasterEqn.java:400)
at jing.rxnSys.ReactionSystem.initializePDepNetwork(ReactionSystem.java:542)
at jing.rxnSys.ReactionModelGenerator.modelGeneration(ReactionModelGenerator.java:1466)
at RMG.main(RMG.java:96)
Exception in thread "main" java.lang.NullPointerException
at jing.rxnSys.Logger.log(Logger.java:160)
at jing.rxnSys.Logger.critical(Logger.java:204)
at RMG.main(RMG.java:106)
I've uploaded fame input and output files at https://github.com/GreenGroup/RMG-Java/downloads
Per discussion with @jwallen, it sounds like this may be related to use of a branch off an outdated version of the master branch...I'll pull in the most recent changes and retry.
I believe we "fixed" this - in that it now falls back to modified strong collision rather than crashing out - in some recent commits, e.g. 61057c47dc456c1efdc5e047454e16ae75062869. You might try that if you're not on an up-to-date branch.
NB. "fixed" is rightly in inverted commas...
On Aug 29, 2011, at 9:48 AM, jwallen wrote:
I believe we "fixed" this - in that it now falls back to modified strong collision rather than crashing out - in some recent commits, e.g. 61057c47dc456c1efdc5e047454e16ae75062869. You might try that if you're not on an up-to-date branch.
Reply to this email directly or view it on GitHub: https://github.com/GreenGroup/RMG-Java/issues/195#issuecomment-1935591
Hi Richard,
Even after updating (i.e. falling back to modified strong collision) one of my job failed. Please find the folder location and error message as below
Folder location:
rajesh@pharos:~/Rajesh/New_jobs_after_Aug_11/MultiT_PM3_pdep_Prun_15000_single_concentration_single_presssure_O2_C_0.15_conversion_restarted_afterupdate$
Error message:
1955 C7H13J(1955) 4.5463E-11 2.2989E-07
ERROR: java.lang.NullPointerException
at jing.rxn.Reaction.getDirection(Reaction.java:924)
at jing.rxnSys.ReactionModelGenerator.writePDepNetworks(ReactionModelGenerator.java:2794)
at jing.rxnSys.ReactionModelGenerator.modelGeneration(ReactionModelGenerator.java:1565)
at RMG.main(RMG.java:96)
This last stack trace looks like issue #207 not #195 to me. Could you paste it in a comment there?