RMG-Py
 RMG-Py copied to clipboard
RMG-Py copied to clipboard
Regarding generation of surface reaction families for Dry Methane Reforming
Topic
General area which your question is related to.
- [ ] Installation of RMG
- Running an RMG job
- [ ] Using RMG API
- [ ] Arkane (formerly CanTherm)
- [ ] Dependencies
- [ ] An error message
Context
I tried to run the example (Dry methane reforming) available from the supplementary paper of Automatic generation of microkinetic mechanisms for heterogeneous catalysis, CF Goldsmith, RH West, J. Phys. Chem. C, 2017, DOI: http://doi.org/10.6084/m9.figshare.4640098.v1
Specifically, I referred to the guide in the jupyter notebook provided, and tried to run the DRM example with the reaction families specified: (Snippet from Jupyter Notebook) --- ch4_co2_seed/input.py 2017-02-08 22:23:42.000000000 -0500 +++ ch4_co2_families/input.py 2017-02-08 22:23:42.000000000 -0500 @@ -1,10 +1,10 @@
Data sources database( thermoLibraries=['surfaceThermo', 'primaryThermoLibrary', 'thermo_DFT_CCSDTF12_BAC','DFT_QCI_thermo'],
- reactionLibraries = [],
- seedMechanisms = ['Deutschmann_Ni_full'], # Full Deutschmann as seed mechanism.
- reactionLibraries = [('Deutschmann_Ni', False)],
- seedMechanisms = [], kineticsDepositories = ['training'],
- kineticsFamilies = 'none', # Turn off all reaction generation.
- kineticsFamilies = 'default', kineticsEstimator = 'rate rules',
Question
After specifying 'default' reaction families in the input.py file, I was not able to replicate the output given in the jupyter notebook: (Snippet from Jupyter Notebook) MODEL GENERATION COMPLETED
The final model core has 49 species and 98 reactions The final model edge has 249 species and 418 reactions
My interest is to understand surface reaction species for heterogeneous catalysts. However, removing "Deutschmann_Ni_full" from the seed mechanism and adding both 'Deutschmann_Ni' in reactionLibraties and 'default' reaction families yield the same result for when I ran "Deutschmann_Ni_full seed mechanism" without any reaction families (i.e., The reaction families only generate new gas-phase reactions, surface reaction species were the same as from the seed mechanisms specific in 'Deutschmann_Ni_full').
I thus decided to add all available surface reaction families:
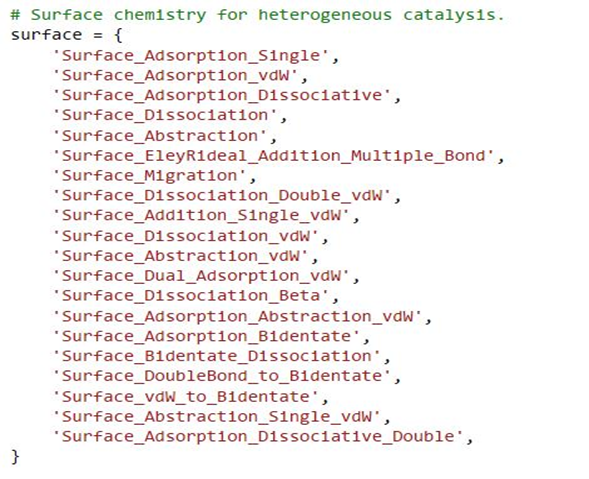 However, the program/ solver just gets stuck after generating a number of species and reactions (after approximately 19 mins; see attached)
RMG Stucked.log
input.txt
However, the program/ solver just gets stuck after generating a number of species and reactions (after approximately 19 mins; see attached)
RMG Stucked.log
input.txt
I have tried to reduce the surface reaction families but cannot replicate the solution derived in the jupyter notebook: MODEL GENERATION COMPLETED
The final model core has 49 species and 98 reactions The final model edge has 249 species and 418 reactions
May I ask the developers/ authors to assist me in this?
Installation Information
Describe your installation method and system information if applicable.
- Windows 10
- Installing RMG in the Linux Subsystem on Windows 10, Installation by Source Using Anaconda Environment for Unix-based Systems: Linux and Mac OSX
- RMG version information:
- RMG-Py: 3.1.0-200-ga20b901ca
- RMG-database: 3.0.0-263-g1618c480b