mdanalysis
 mdanalysis copied to clipboard
mdanalysis copied to clipboard
[DOC]: Updated references to BibTex format
Fixes #3766 and entirely fixes #2837
Changes made in this Pull Request:
- Updated all references in
references.rstto BibTex format. - Added references to
references.bib. - Fixed typos.
- Fixed links.
PR Checklist
- N/A Tests
- [x] Docs
- N/A CHANGELOG updated
- [x] Issue raised/referenced
Comments:
This time I added changes split into different small commits, which hopefully will help reviewers to identify the changes in each section of the references.rst.
I will leave some specific comments in the code to double-check check I didn't miss anything and that we are not losing information.
Hello @ojeda-e! Thanks for updating this PR. We checked the lines you've touched for PEP 8 issues, and found:
- In the file
package/MDAnalysis/analysis/helix_analysis.py:
Line 42:80: E501 line too long (120 > 79 characters)
Comment last updated at 2022-08-28 17:13:45 UTC
Codecov Report
Patch and project coverage have no change.
Comparison is base (
66a2b21) 93.62% compared to head (4dcf2b3) 93.62%.
Additional details and impacted files
@@ Coverage Diff @@
## develop #3775 +/- ##
========================================
Coverage 93.62% 93.62%
========================================
Files 193 193
Lines 25295 25295
Branches 4063 4063
========================================
Hits 23683 23683
Misses 1096 1096
Partials 516 516
| Files Changed | Coverage Δ | |
|---|---|---|
| package/MDAnalysis/analysis/align.py | 96.38% <ø> (ø) |
|
| package/MDAnalysis/analysis/dielectric.py | 100.00% <ø> (ø) |
|
| package/MDAnalysis/analysis/diffusionmap.py | 100.00% <ø> (ø) |
|
| package/MDAnalysis/analysis/encore/similarity.py | 85.62% <ø> (ø) |
|
| package/MDAnalysis/analysis/helix_analysis.py | 98.65% <ø> (ø) |
|
| ...DAnalysis/analysis/hydrogenbonds/hbond_analysis.py | 97.20% <ø> (ø) |
|
| package/MDAnalysis/analysis/leaflet.py | 94.93% <ø> (ø) |
|
| package/MDAnalysis/analysis/msd.py | 100.00% <ø> (ø) |
|
| package/MDAnalysis/analysis/pca.py | 99.49% <ø> (ø) |
|
| package/MDAnalysis/analysis/rms.py | 93.02% <ø> (ø) |
|
| ... and 4 more |
:umbrella: View full report in Codecov by Sentry.
:loudspeaker: Have feedback on the report? Share it here.
Thanks for all the comments @orbeckst, I'll address them in the next push, but also I think I may need some extra time here because the docs are not building properly and I can't identify the root of the issue. It is either (something I introduced in) package/MDAnalysis/analysis/rms.py, or package/MDAnalysis/analysis/align.py, or package/MDAnalysis/lib/qcprot.pyx. Something there is making docs break in the references page and similarly, in align. Weirdly enough, docs build properly locally. I'll check what's going on.
@orbeckst I addressed all your comments, thanks for pointing out the missing reference and the inconsistency of the dois in the bib references.
I found that updating references in package/MDAnalysis/lib/qcprot.pyx is problematic since it is breaking docs (see outdated references page ). It isn't clear to me why it fails here. Docs in .pyx files is supported. For now, I restored the previous version (old format for two references) to build docs properly. @MDAnalysis/docteam: Suggestions on how to update the refs to BibTex format without breaking docs are welcome.
Not sure if it's best to open a separate issue for the missing references in qcprot.pyx or try to fix it in this PR (with some extra help). If we choose the first one, this PR is ready.
Looks awful when it breaks :-p.
Did you successfully include bibtex citations from any other pyx files?
Did you try removing citations piece by piece to see if it's any specific one?
Looks awful when it breaks :-p.
Agreed!
Did you successfully include bibtex citations from any other pyx files?
Not really. The citations I found in other pyx files are restricted to the header that reads "# Please cite your use of MDAnalysis in published work: (...)" , such as
-
or even nsgrid.pyx that mentions "Frenkel and Smit" but has no actual reference.
The only .pyx file with references to update was qcprot.pyx in package/MDAnalysis/lib/qcprot.pyx, references Theobald2005 and Liu2010.
Did you try removing citations piece by piece to see if it's any specific one?
Yes, I did. Both Theobald2005 and Liu2010 in qcprot.pyx. I tried to add them separately, one by one. No matter which one I add, the docs break as soon as there is BibTex format on it.
Are there any intermediate files that are created? If so, do the references look correct in the intermediate files?
The issue about apidoc and pyx files https://github.com/sphinx-doc/sphinx/issues/1117 only addresses sphinx itself but does not mention bibtex. Is there any indication that the bibtex part might break??
Are there any intermediate files that are created? If so, do the references look correct in the intermediate files?
Not that I am aware of. The error shows a wrong indentation very far up from the line I edited. The reference was added in line 90 (see commit) while the error suggests (see step 5 of the CI) a wrong indentation in line 12-13:
docstring of MDAnalysis.lib.qcprot:12: ERROR: Unexpected indentation.
docstring of MDAnalysis.lib.qcprot:13: WARNING: Block quote ends without a blank line; unexpected unindent.
Please note that the error shows that the error is triggered when only one reference in BibTex is added.
The issue about apidoc and pyx files sphinx-doc/sphinx#1117 only addresses sphinx itself but does not mention bibtex. Is there any indication that the bibtex part might break??
Unfortunately, I am failing to find information on the topic. Since the first time docs break, I tried to dig in but all the info I find related to support is related to Sphinx only (as the one you pointed out), not Sphinx + sphinxcontrib-bibtex extension.
I sent a message to the Sphinx mailing list asking for information (since it is a extension, not a problem on Sphinx itself). I also opened an issue in sphinxcontrib-bibtex asking if cython files are supported. See sphinxcontrib-bibtex/issues#308.
After checking that the BibTex extension is supported for Cython files, I tried several approaches to solve the problem but I failed to find a solution. From what I can see, the problem is in the Theobald reference, which happened to be in a Cython file as well. Adding the reference to the bibliography list breaks the HTML when adding it to diffusionmap.py or references.rst, and also happens when added in the cython file, qcprot.pyx. Honestly, I can't figure out what is the reason behind it despite trying several options. This is getting very complicated to debug because despite having identical versions for all sphinx packages and associated extensions, docutils, and other relevant packages, what I build locally is dramatically different from what is generated in the CI.
Example: snapshots of MDAnalysis.analysis.rms
local build
 vs.
CI build
vs.
CI build
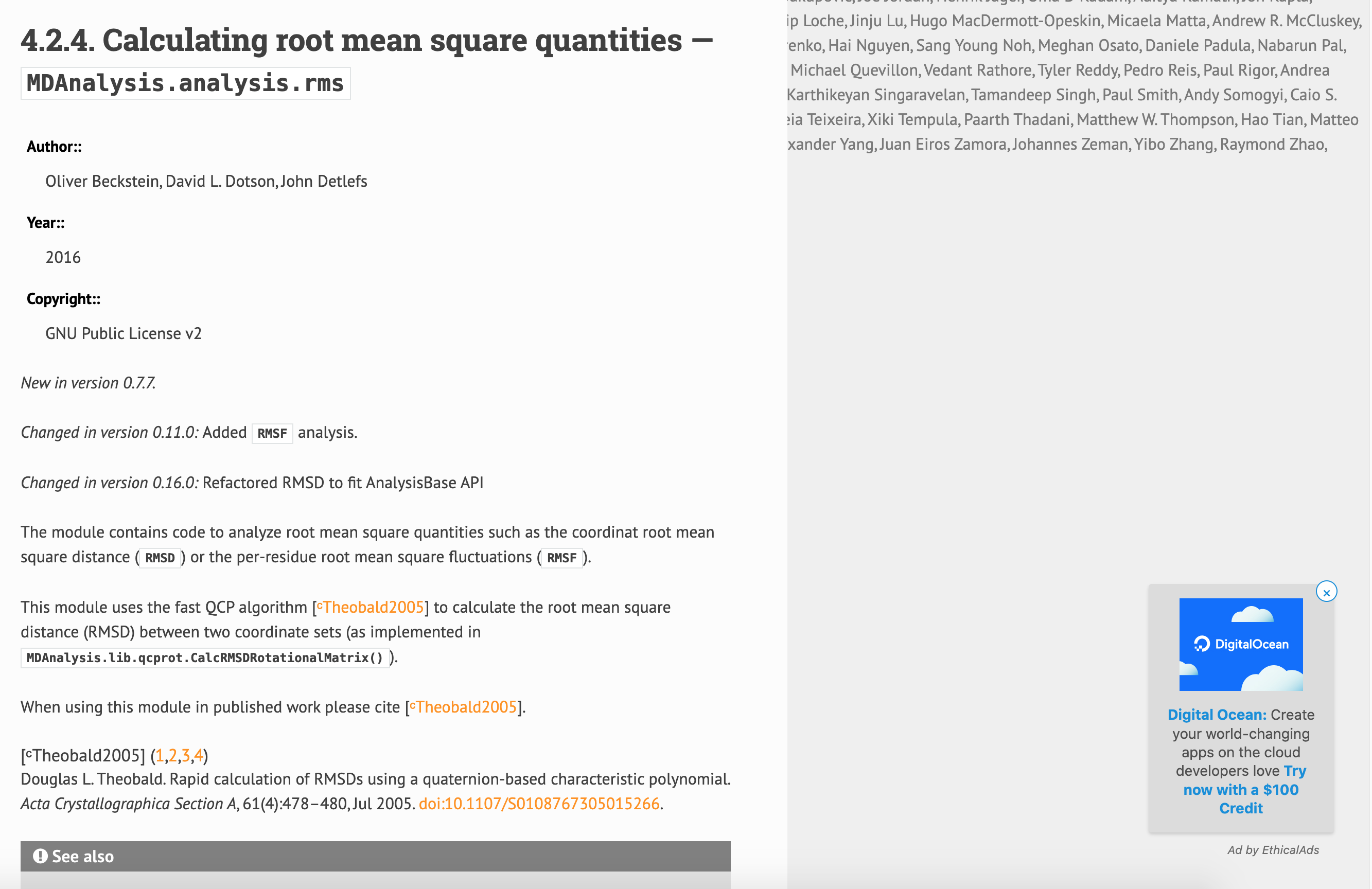
Additionally, building locally doesn't trigger errors than are caught by the CI.
I am kind of giving up here because no matter what I try the docs break. I tried doing a diff between the in rms.py HTML files generated locally and the CI and the output is more than I'd expect. Here is a snapshot, where the one on the left is generated by the CI and the one on the right is generated locally.
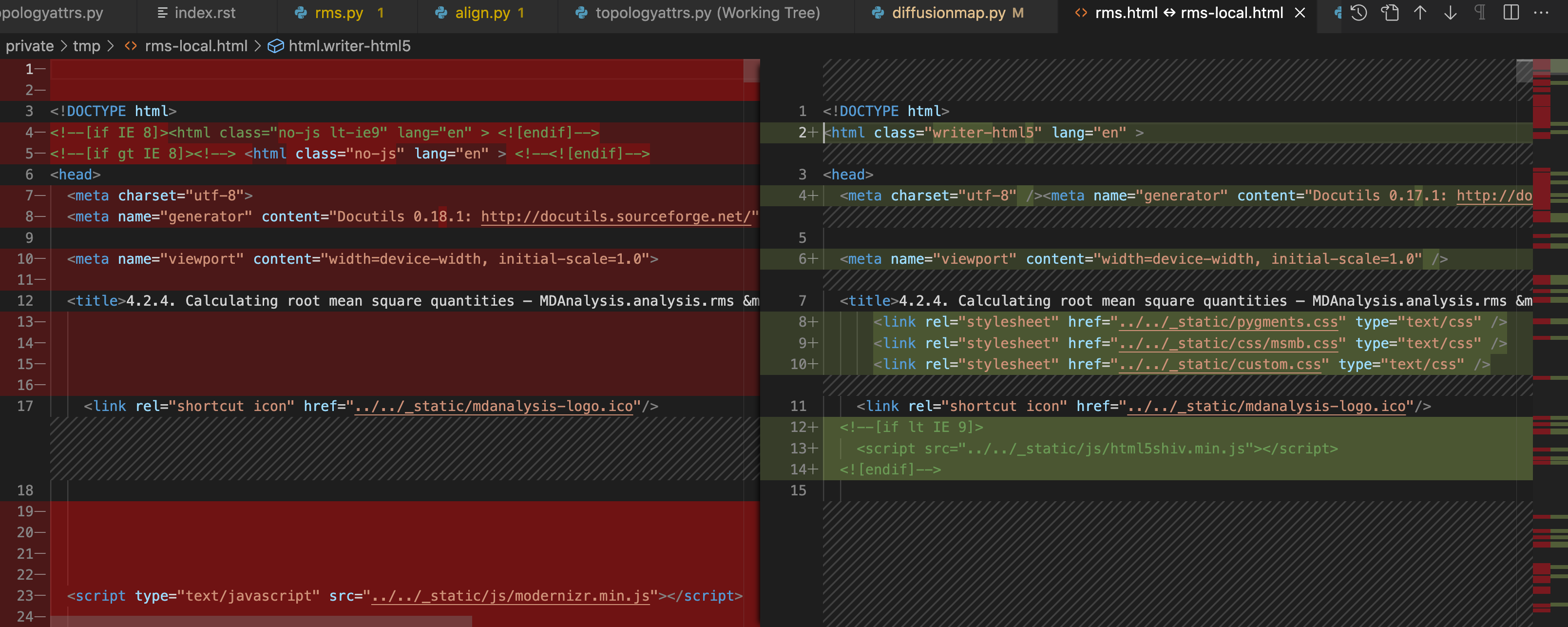 The one from the CI is also unexpected in the version of docutils. I would expect docutils==0.17.1, as shown in during building, not 0.18.1, as seems to be the one used to build docs (BTW, if someone can explain to me why is there a difference, I'd appreciate it).
The one from the CI is also unexpected in the version of docutils. I would expect docutils==0.17.1, as shown in during building, not 0.18.1, as seems to be the one used to build docs (BTW, if someone can explain to me why is there a difference, I'd appreciate it).
In summary, I don't know how to proceed here. I'm tempted to put Theobald references back to rst and have something functional that doesn't break the three problematic pages: package/MDAnalysis/analysis/rms.py package/MDAnalysis/analysis/align.py, qcprot.pyxand references.rst.
Any suggestions are more than welcome.
Thank you for digging deeply into the bibtex extension and checking with upstream. All of this is really important when we adopt a new technology — we want to understand problems at the beginning.
Just so that I understand what's happening — please correct me:
- You narrowed down problems to the Theobald reference that is used in a pyx file as well as other files. If you don't have it in the pyx then no problems appear.
- Building the docs locally works and they appear as expected.
- The builds that are displayed on RTD as part of the PR build differ visually from the local ones.
- "Additionally, building locally doesn't trigger errors than are caught by the CI." — what are the errors that you refer to? Did you mean the "docstring of MDAnalysis.lib.qcprot:12: ERROR: Unexpected indentation."?
Concerning (3), what is the difference that you consider incorrect?
(I am asking because as far as I can tell, RTD doc formatting always differs slightly between our published docs and the RTD version, see for example rms module in PR 3799 vs rms module docs v2.2.)
About (4): Having sphinx throw errors is annoying as it stops tests. Finding a fix for it would be important. Perhaps adding additional text afterwards, checking spaces vs tabs, ... and other Voodo-coding might help? (... it's super-annoying, I know ...)
Perhaps the fact that the RTD html has class="no-js" and ours has class="writer-html5" is significant. I don't know. However, as long as the RTD version for your PR looks similar to the RTD version on other PRs and you confirmed locally that it builds correctly and sphinx does not throw errors, then we can chance it and make the changes and see what it looks like in production devel docs.
Maybe @lilyminium has some more insights?
@ojeda-e did I understand your findings correctly?
Any idea what else to try or how to solve this problem, maybe with a work-around?
Sorry for the late reply @orbeckst. For some reason, Github hasn't sent me activity notifications for a while.
Just so that I understand what's happening — please correct me:
- You narrowed down problems to the Theobald reference that is used in a pyx file as well as other files. If you don't > have it in the pyx then no problems appear.
Correct.
- Building the docs locally works and they appear as expected.
- The builds that are displayed on RTD as part of the PR build differ visually from the local ones.
Correct.
- "Additionally, building locally doesn't trigger errors than are caught by the CI." — what are the errors that you refer to? Did you mean the "docstring of MDAnalysis.lib.qcprot:12: ERROR: Unexpected indentation."?
Yes, exactly that.
(I am asking because as far as I can tell, RTD doc formatting always differs slightly between our published docs and the RTD version, see for example rms module in PR 3799 vs rms module docs v2.2.)
Oh. I didn't know that was a thing. I really appreciate you pointing that out. Was debugging step by step to find out why the rendered docs looked like that. The same happens in the docs here updated, so it won't probably be an issue.
If there is a chance that the docs may not render properly, I could offer a solution. Considering we have Theobald2005 in several files:
./doc/sphinx/source/documentation_pages/references.rst./MDAnalysis/analysis/align.py./MDAnalysis/analysis/diffusionmap.py./MDAnalysis/analysis/rms.py./MDAnalysis/lib/qcprot.pyx
Locally, the docs render as expected using the following prefixes for different files:
| file | prefix |
|---|---|
| references.rst | ᵃ |
| align.py | ᵇ |
| diffusionmap.py | - |
| rms.py | ᵈ |
| qcprot.pyx | ᶜ |
(Other than the one for references.rst ᵃ, because I wanted consistency on that page, the prefixes were assigned randomly; - means no prefix, equivalent to an empty prefix.)
With this solution, the references.rst page, where Theobald shows up would look like this (rendered locally):
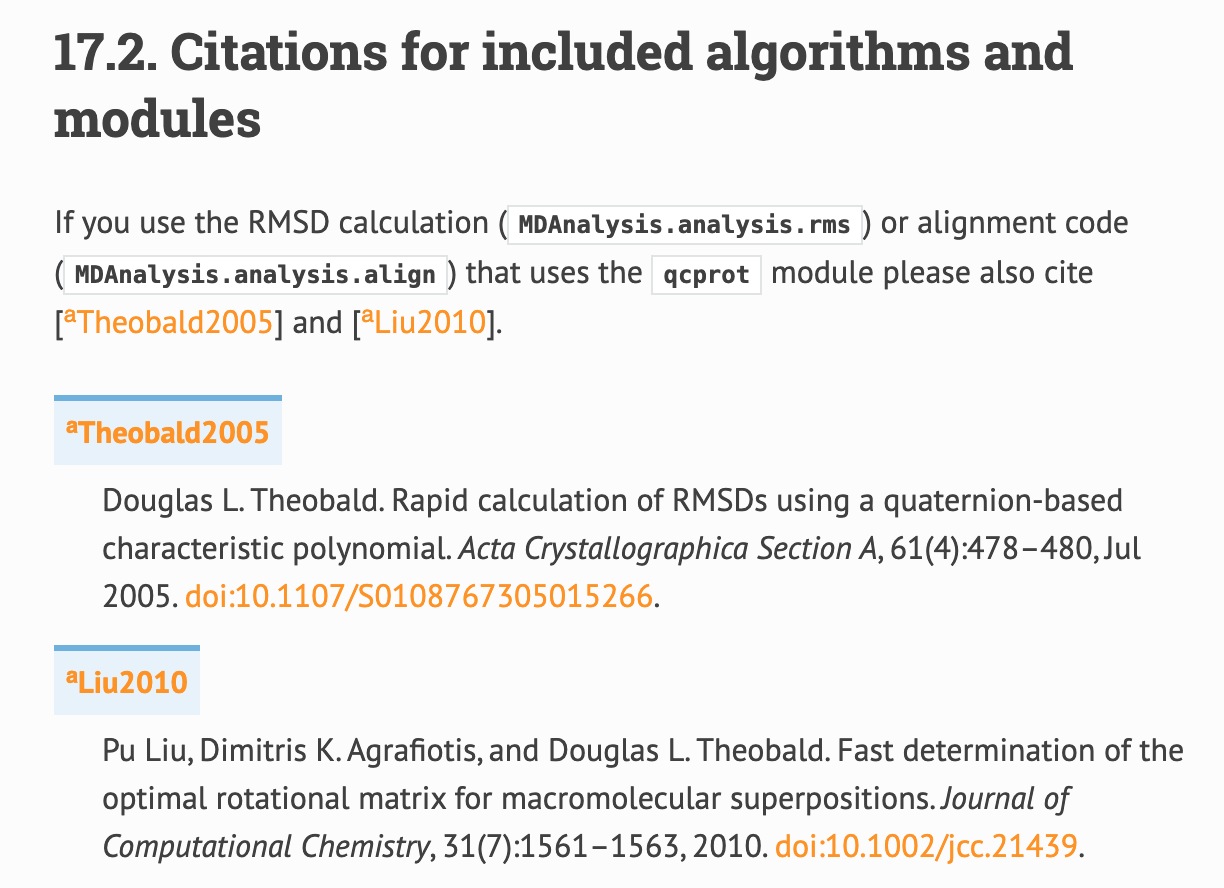
This solution renders locally for all the other files above listed. The only thing is that this will likely look different on the CI.
Would it be too risky or complicated to create something like a test issue where I only change Theobald, merge it, and check how does it look like and if it's too bad we undo the merge?
@ojeda-e my turn to apologize... sorry, lost track of the PR.
If you have a solution that renders the reference and does not crash CI/RTD then I am happy with it.
I looked at https://mdanalysis--3775.org.readthedocs.build/en/3775/documentation_pages/references.html#citations-for-included-algorithms-and-modules and the list of references for included algorithms looks like a mess, as you indicated. If it only looks bad on RTD but ok in our docs https://docs.mdanalysis.org then I'd be ok with it. I don't know if e.g. @lilyminium has a different opinion.
Would it be too risky or complicated to create something like a test issue where I only change Theobald, merge it, and check how does it look like and if it's too bad we undo the merge?
Do you want to do a single change and then see how it shows up at https://docs.mdanalysis.org to avoid looking at it through the lens of RTD?
If yes, then please go ahead and create that PR. If the other one does what we expect (looks ok in our docs, broken references page on RTD) then we can merge develop into this PR and ultimately merge this one, too.
docstring of MDAnalysis.lib.qcprot:12: ERROR: Unexpected indentation
Hmm, yes, I'd say go ahead and see the change works but maybe raise a new (low-priority) issue to check out what's going on with CI... maybe the docs dependency versions need updating or something similar.
If you want to do a dry-run on a fork, you can change CI to build to a github-pages URL of your own. I did that here: https://mdadocs.minium.com.au/2.4.0-dev0/documentation_pages/references.html#citations-for-included-algorithms-and-modules is current for commit 79d4feba2dbc0adbbe2bfd21cd0bfa33892510ce
(it's the full docs so you can click around, but every now-and-then you might hit a redirect page that sends you to docs.mdanalysis.org)
There was some discussion of the need for pyyaml in #3842 https://github.com/MDAnalysis/mdanalysis/pull/3842/files#r1027222362
Not sure if relevant.
@ojeda-e @orbeckst is this still PR still active?
In principle, these changes still need to be done to change references, as far as I can tell.
I merged develop into this branch and maybe citations become magically correct?? (Hope springs eternal...)
Linter Bot Results:
Hi @ojeda-e! Thanks for making this PR. We linted your code and found the following:
Some issues were found with the formatting of your code.
| Code Location | Outcome |
|---|---|
| main package | ⚠️ Possible failure |
| testsuite | ✅ Passed |
Please have a look at the darker-main-code and darker-test-code steps here for more details: https://github.com/MDAnalysis/mdanalysis/actions/runs/5721087324/job/15502129369
Please note: The black linter is purely informational, you can safely ignore these outcomes if there are no flake8 failures!
@lilyminium from the conflicts I see that footblibliography appears in place of the explicit footnotes from this PR. Does this mean that this PR is no longer needed?
Yes and no -- @ojeda-e put a lot of amazing work into fixing up all our references and making sure everything was neatly bibtex that would be a great pity to lose, and I only went through and fixed existing, previously merged bibliographies when I updated the theme. Presumably the rest of the 18 files with changes that don't have conflicts are all improvements of changing footnotes to bibtex references. I'm not sure how much @ojeda-e wants to keep working on this, but it's been quite a while -- maybe the easiest way to resolve this PR would be for me to either take over and resolve the conflicts/convert things into local bibliographies, or cherry-pick commits into a different PR and go from there.
Hi @lilyminium, please feel free to cherry pick anything is useful for you and close this PR if that's the best approach for the organization to move forward. Thanks for acknowledging the work, I appreciate it.