cadquery
 cadquery copied to clipboard
cadquery copied to clipboard
3MF File support
I added basic support for the 3MF file format. It works similar to the already supported AMF format. Just like AMF, we do in no way leverage the full potential of the format quite yet.
In future, we might want to look into using these formats for Assemblies. Maybe also look into some of the more advanced geometrical features, which would require custom tessellation.
The generated files were tested with PrusaSlicer and Cura.
Codecov Report
Merging #1105 (114e4cc) into master (d4cdeeb) will increase coverage by
0.08%. The diff coverage is100.00%.
@@ Coverage Diff @@
## master #1105 +/- ##
==========================================
+ Coverage 93.90% 93.98% +0.08%
==========================================
Files 25 26 +1
Lines 5328 5404 +76
Branches 979 993 +14
==========================================
+ Hits 5003 5079 +76
Misses 193 193
Partials 132 132
| Impacted Files | Coverage Δ | |
|---|---|---|
| cadquery/occ_impl/exporters/__init__.py | 92.43% <100.00%> (+0.61%) |
:arrow_up: |
| cadquery/occ_impl/exporters/threemf.py | 100.00% <100.00%> (ø) |
:mega: We’re building smart automated test selection to slash your CI/CD build times. Learn more
I'm struggling to fully introduce the pyecma376-2 dependency. Suggestions from anyone familiar with the build-pipeline are welcome.
@SDI8 You can try adding the dependency to meta.yml as well. https://github.com/CadQuery/cadquery/blob/master/conda/meta.yaml
@jmwright Thank you! Meta.xml requirements are obviously conda packages. But since I couldn't find any suitable package capable of doing OPC/OpenXML/ecma376-2 on conda, I had to go with pypi.
I should have gone without the pyecma dependency from the beginning. I removed it and it hardly made a difference in complexity :+1:
Just for the record: 3MF supports compounds (ie. groups of objects). I have implemented that and it works really nicely in Cura. It shows the objects as a "group", and allows you to manipulate them in unison or individually. Other applications load the file well, too! Prusa Slicer, however, just rejects to import those files. In general i'm a bit underwhelmed by the 3MF support in Prusa Slicer - in practice it only works with the files it generated itself. ~~I'll leave this PR as it is, as support for PrusaSlicer is likely going to be important for some. However, preserving the hierarchy of shapes would be really nice when using 3MF for assemblies - somewhere in the future.~~
EDIT: I got it working, Prusa is very specific about the IDs of objects.
@SDI8 Where does this PR stand? Is it ready for review? I'm assuming this means zlib will need to be added to the dependencies in the various config files, correct?
@SDI8 Where does this PR stand? Is it ready for review? I'm assuming this means zlib will need to be added to the dependencies in the various config files, correct?
Yes, the PR is ready for review. Zlib should be builtin and always available in Python. The exception are some rare, custom builds. Therefore we do not require an explicit dependency.
@SDI8 Thank you for this, and thanks for adding information to the documentation related to 3MF support so that it doesn't have to be done separately.
I checked out out this branch, created a 10x10x10 box, exported it to 3MF, then opened it in PrusaSlicer. It opened, but I got an error that there are "24 open edges". When I import it into Cura 4.4.1 (I rarely use Cura), the model is placed at the lower left corner of the bed and Cura complains that it can't slice it. Once I move the model to the center of the bed it works and slices fine. I'm not sure why the origin is set that way with Cura. PrusaSlicer puts the model directly in the center of the bed.
Here is the code that I used to make the box and export it. I did it from the Python REPL. I had set up a Python virtual environment with the cadquery-ocp package installed from PyPi.
import cadquery as cq
result = cq.Workplane().box(10, 10, 10)
cq.exporters.export(result, "/home/jwright/Downloads/out3.3mf")
Here's a screenshot of the PrusaSlicer error.
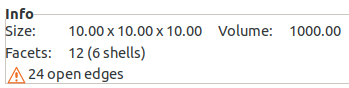
@jmwright thanks for your feedback. Yes, Cura places 3MF files at origin (0,0). In contrast to other formats, it does not perform the auto-arrange on import. And it will complain when models are on the edge of the build-plate.
The open edges in Prusa is a problem with the AMF writer, too. Which does not surprise me, as the formats are very similar in that regard. The model is watertight, but it must be down to the way the result from the tessellation is arranged. I might have a look at that.
Yes, the PR is ready for review. Zlib should be builtin and always available in Python. The exception are some rare, custom builds. Therefore we do not require an explicit dependency.
LGTM, why do we need to support "rare, custom builds" though? Zlib is in the standard library of Python.
@adam-urbanczyk Feel free to merge once the discussion about zlib is concluded.
@adam-urbanczyk Any objection to me merging this?
@SDI8 can you comment on the need for custom Python build support?
@SDI8 can you comment on the need for custom Python build support?
That's up to you. It might be a very rare case, but it's only a few extra lines, which guard against an error which would be hard to solve for the user.
OK, let's leave it for now. I general though, I'd propose to not handle such special cases unless they are somewhat common.
Thanks for this very nice contribution @SDI8 !